Please be patient as the page loads
|
PYGO1_MOUSE
|
||||||
| SwissProt Accessions | Q9D0P5 | Gene names | Pygo1 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PYGO1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.971384 (rank : 2) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y3Y4 | Gene names | PYGO1 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 1. | |||||
|
PYGO1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9D0P5 | Gene names | Pygo1 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 1. | |||||
|
PYGO2_HUMAN
|
||||||
| θ value | 1.7238e-59 (rank : 3) | NC score | 0.793713 (rank : 3) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BRQ0, Q8WYZ4, Q96CY2 | Gene names | PYGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 2. | |||||
|
GAB2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 4) | NC score | 0.080479 (rank : 9) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 5) | NC score | 0.043687 (rank : 29) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 6) | NC score | 0.078060 (rank : 10) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 7) | NC score | 0.076630 (rank : 12) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
NR1D1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 8) | NC score | 0.025711 (rank : 47) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20393, Q15304 | Gene names | NR1D1, EAR1, HREV, THRAL | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D1 (V-erbA-related protein EAR-1) (Rev- erbA-alpha). | |||||
|
ING3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 9) | NC score | 0.072307 (rank : 15) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NXR8, O60394, Q6GMT3, Q7Z762, Q969G0, Q96DT4, Q9HC99, Q9P081 | Gene names | ING3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.043113 (rank : 30) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.039558 (rank : 34) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
PHF14_MOUSE
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.074687 (rank : 13) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.058274 (rank : 18) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.043800 (rank : 28) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.073586 (rank : 14) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
PTN14_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.020716 (rank : 50) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.017607 (rank : 55) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
JAD1A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.048784 (rank : 27) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
PHF14_HUMAN
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.070725 (rank : 16) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
SF01_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.056772 (rank : 21) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
SF01_MOUSE
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.058111 (rank : 19) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
SPT5H_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.093277 (rank : 5) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
SPT5H_MOUSE
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.093535 (rank : 4) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
HELZ_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.048838 (rank : 26) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
LATS1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.008281 (rank : 66) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
TAB2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.040704 (rank : 32) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYJ8, O94838, Q6I9W8, Q76N06, Q9UFP7 | Gene names | MAP3K7IP2, KIAA0733, TAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.077584 (rank : 11) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
ORC2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.038034 (rank : 37) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13416, Q13204 | Gene names | ORC2L, ORC2 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 2. | |||||
|
TAB3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.059078 (rank : 17) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
ZN645_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.086031 (rank : 6) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N7E2, Q6DJY9 | Gene names | ZNF645 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 645. | |||||
|
SN1L2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.004043 (rank : 69) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0K1, O94878, Q6AZE2, Q76N03, Q8NCV7, Q96CZ8 | Gene names | SNF1LK2, KIAA0781, QIK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Qin- induced kinase). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.083857 (rank : 8) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.034815 (rank : 41) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
ING3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.042502 (rank : 31) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VEK6, Q99JS6, Q9ERB2 | Gene names | Ing3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.038170 (rank : 36) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.028962 (rank : 44) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
SIP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.005354 (rank : 67) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0G7 | Gene names | Zfhx1b, Sip1, Zfx1b | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 1b (Smad-interacting protein 1). | |||||
|
WASL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.039804 (rank : 33) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
ZMYM3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.023784 (rank : 48) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
DAAM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.016694 (rank : 58) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86T65, Q5T4U0, Q9NQI5, Q9Y4G0 | Gene names | DAAM2, KIAA0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.018336 (rank : 54) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
F113A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.025998 (rank : 46) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P1Z5 | Gene names | Fam113a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM113A. | |||||
|
NUFP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.037097 (rank : 40) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXX8, Q9CV69 | Gene names | Nufip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.037820 (rank : 38) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SIP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.005241 (rank : 68) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60315, Q9UED1 | Gene names | ZFHX1B, KIAA0569, SIP1, ZFX1B | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 1b (Smad-interacting protein 1) (SMADIP1). | |||||
|
CCNL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.013764 (rank : 60) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q52KE7, Q8BQ75, Q8R5H9, Q922K0, Q9CSZ3, Q9WV44 | Gene names | Ccnl1, Ania6a, Ccn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L) (Cyclin Ania-6a). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.057470 (rank : 20) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.055857 (rank : 23) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PARC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.013007 (rank : 61) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.056102 (rank : 22) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.020482 (rank : 51) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
HNRL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.016507 (rank : 59) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
NOP56_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.016716 (rank : 57) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D6Z1, Q3UD45, Q8BVL1, Q8VDT2, Q99LT8, Q9CT15 | Gene names | Nol5a, Nop56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
PCD15_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.003858 (rank : 70) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PJ1 | Gene names | Pcdh15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
PRPC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.085249 (rank : 7) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.012596 (rank : 62) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
SMN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.039481 (rank : 35) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16637, Q13119, Q96J51 | Gene names | SMN1, SMN, SMNT | |||
|
Domain Architecture |
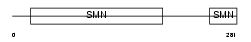
|
|||||
| Description | Survival motor neuron protein (Component of gems 1) (Gemin-1). | |||||
|
CASC5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.012481 (rank : 63) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NG31, Q8NHE1, Q8WXA6, Q9HCK2, Q9NR92 | Gene names | CASC5, KIAA1570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein) (ALL1- fused gene from chromosome 15q14) (AF15q14) (D40/AF15q14 protein). | |||||
|
ENAM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.023780 (rank : 49) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
H6ST2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.010745 (rank : 65) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96MM7, Q4VC07, Q6PIC4, Q86SM9, Q8N3T4, Q8NBN4, Q96SJ4 | Gene names | HS6ST2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 2 (EC 2.8.2.-) (HS6ST-2). | |||||
|
MAK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.000842 (rank : 71) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04859 | Gene names | Mak, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MAK (EC 2.7.11.22) (Male germ cell- associated kinase) (Protein kinase RCK). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.037312 (rank : 39) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
PHF20_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.031351 (rank : 42) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.030484 (rank : 43) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.028359 (rank : 45) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RXRB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.011095 (rank : 64) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P28704, P33243 | Gene names | Rxrb, Nr2b2 | |||
|
Domain Architecture |
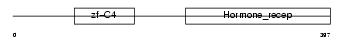
|
|||||
| Description | Retinoic acid receptor RXR-beta (Retinoid X receptor beta) (MHC class I regulatory element-binding protein H-2RIIBP). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.016787 (rank : 56) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SSBP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.018882 (rank : 52) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BWW4, Q9BTM0, Q9BWW3 | Gene names | SSBP3, SSDP, SSDP1 | |||
|
Domain Architecture |
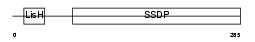
|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein). | |||||
|
SSBP3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.018882 (rank : 53) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D032, Q99LC6, Q9EQP3 | Gene names | Ssbp3, Last, Ssdp1 | |||
|
Domain Architecture |
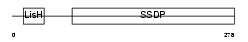
|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein) (Lck-associated signal transducer). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.050362 (rank : 25) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.052002 (rank : 24) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
PYGO1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9D0P5 | Gene names | Pygo1 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 1. | |||||
|
PYGO1_HUMAN
|
||||||
| NC score | 0.971384 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y3Y4 | Gene names | PYGO1 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 1. | |||||
|
PYGO2_HUMAN
|
||||||
| NC score | 0.793713 (rank : 3) | θ value | 1.7238e-59 (rank : 3) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BRQ0, Q8WYZ4, Q96CY2 | Gene names | PYGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 2. | |||||
|
SPT5H_MOUSE
|
||||||
| NC score | 0.093535 (rank : 4) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
SPT5H_HUMAN
|
||||||
| NC score | 0.093277 (rank : 5) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
ZN645_HUMAN
|
||||||
| NC score | 0.086031 (rank : 6) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N7E2, Q6DJY9 | Gene names | ZNF645 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 645. | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.085249 (rank : 7) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.083857 (rank : 8) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
GAB2_HUMAN
|
||||||
| NC score | 0.080479 (rank : 9) | θ value | 0.0563607 (rank : 4) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UQC2, O60317 | Gene names | GAB2, KIAA0571 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (pp100). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.078060 (rank : 10) | θ value | 0.0563607 (rank : 6) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.077584 (rank : 11) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.076630 (rank : 12) | θ value | 0.0736092 (rank : 7) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
PHF14_MOUSE
|
||||||
| NC score | 0.074687 (rank : 13) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.073586 (rank : 14) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
ING3_HUMAN
|
||||||
| NC score | 0.072307 (rank : 15) | θ value | 0.21417 (rank : 9) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NXR8, O60394, Q6GMT3, Q7Z762, Q969G0, Q96DT4, Q9HC99, Q9P081 | Gene names | ING3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.070725 (rank : 16) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
TAB3_HUMAN
|
||||||
| NC score | 0.059078 (rank : 17) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N5C8, Q6VQR0 | Gene names | MAP3K7IP3, TAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3) (NF-kappa-B-activating protein 1). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.058274 (rank : 18) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
SF01_MOUSE
|
||||||
| NC score | 0.058111 (rank : 19) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64213, O08817, P70167, Q61454, Q921Z4 | Gene names | Sf1, Zfm1, Zfp162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (mZFM) (Zinc finger gene in MEN1 locus) (Mammalian branch point- binding protein mBBP) (BBP) (CW17). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.057470 (rank : 20) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
SF01_HUMAN
|
||||||
| NC score | 0.056772 (rank : 21) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.056102 (rank : 22) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.055857 (rank : 23) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.052002 (rank : 24) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.050362 (rank : 25) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
HELZ_HUMAN
|
||||||
| NC score | 0.048838 (rank : 26) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
JAD1A_HUMAN
|
||||||
| NC score | 0.048784 (rank : 27) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.043800 (rank : 28) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.043687 (rank : 29) | θ value | 0.0563607 (rank : 5) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.043113 (rank : 30) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
ING3_MOUSE
|
||||||
| NC score | 0.042502 (rank : 31) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VEK6, Q99JS6, Q9ERB2 | Gene names | Ing3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
TAB2_HUMAN
|
||||||
| NC score | 0.040704 (rank : 32) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYJ8, O94838, Q6I9W8, Q76N06, Q9UFP7 | Gene names | MAP3K7IP2, KIAA0733, TAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2). | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.039804 (rank : 33) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.039558 (rank : 34) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
SMN_HUMAN
|
||||||
| NC score | 0.039481 (rank : 35) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16637, Q13119, Q96J51 | Gene names | SMN1, SMN, SMNT | |||
|
Domain Architecture |

|
|||||
| Description | Survival motor neuron protein (Component of gems 1) (Gemin-1). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.038170 (rank : 36) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
ORC2_HUMAN
|
||||||
| NC score | 0.038034 (rank : 37) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13416, Q13204 | Gene names | ORC2L, ORC2 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 2. | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.037820 (rank : 38) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.037312 (rank : 39) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NUFP1_MOUSE
|
||||||
| NC score | 0.037097 (rank : 40) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXX8, Q9CV69 | Gene names | Nufip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.034815 (rank : 41) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
PHF20_HUMAN
|
||||||
| NC score | 0.031351 (rank : 42) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.030484 (rank : 43) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.028962 (rank : 44) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.028359 (rank : 45) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
F113A_MOUSE
|
||||||
| NC score | 0.025998 (rank : 46) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P1Z5 | Gene names | Fam113a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM113A. | |||||
|
NR1D1_HUMAN
|
||||||
| NC score | 0.025711 (rank : 47) | θ value | 0.0961366 (rank : 8) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20393, Q15304 | Gene names | NR1D1, EAR1, HREV, THRAL | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D1 (V-erbA-related protein EAR-1) (Rev- erbA-alpha). | |||||
|
ZMYM3_HUMAN
|
||||||
| NC score | 0.023784 (rank : 48) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
ENAM_HUMAN
|
||||||
| NC score | 0.023780 (rank : 49) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
PTN14_HUMAN
|
||||||
| NC score | 0.020716 (rank : 50) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15678 | Gene names | PTPN14, PEZ | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 14 (EC 3.1.3.48) (Protein-tyrosine phosphatase pez). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.020482 (rank : 51) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
SSBP3_HUMAN
|
||||||
| NC score | 0.018882 (rank : 52) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BWW4, Q9BTM0, Q9BWW3 | Gene names | SSBP3, SSDP, SSDP1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein). | |||||
|
SSBP3_MOUSE
|
||||||
| NC score | 0.018882 (rank : 53) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D032, Q99LC6, Q9EQP3 | Gene names | Ssbp3, Last, Ssdp1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein) (Lck-associated signal transducer). | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.018336 (rank : 54) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.017607 (rank : 55) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.016787 (rank : 56) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
NOP56_MOUSE
|
||||||
| NC score | 0.016716 (rank : 57) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D6Z1, Q3UD45, Q8BVL1, Q8VDT2, Q99LT8, Q9CT15 | Gene names | Nol5a, Nop56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
DAAM2_HUMAN
|
||||||
| NC score | 0.016694 (rank : 58) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86T65, Q5T4U0, Q9NQI5, Q9Y4G0 | Gene names | DAAM2, KIAA0381 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 2. | |||||
|
HNRL1_MOUSE
|
||||||
| NC score | 0.016507 (rank : 59) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
CCNL1_MOUSE
|
||||||
| NC score | 0.013764 (rank : 60) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q52KE7, Q8BQ75, Q8R5H9, Q922K0, Q9CSZ3, Q9WV44 | Gene names | Ccnl1, Ania6a, Ccn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L) (Cyclin Ania-6a). | |||||
|
PARC_HUMAN
|
||||||
| NC score | 0.013007 (rank : 61) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.012596 (rank : 62) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
CASC5_HUMAN
|
||||||
| NC score | 0.012481 (rank : 63) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NG31, Q8NHE1, Q8WXA6, Q9HCK2, Q9NR92 | Gene names | CASC5, KIAA1570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein) (ALL1- fused gene from chromosome 15q14) (AF15q14) (D40/AF15q14 protein). | |||||
|
RXRB_MOUSE
|
||||||
| NC score | 0.011095 (rank : 64) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P28704, P33243 | Gene names | Rxrb, Nr2b2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-beta (Retinoid X receptor beta) (MHC class I regulatory element-binding protein H-2RIIBP). | |||||
|
H6ST2_HUMAN
|
||||||
| NC score | 0.010745 (rank : 65) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96MM7, Q4VC07, Q6PIC4, Q86SM9, Q8N3T4, Q8NBN4, Q96SJ4 | Gene names | HS6ST2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 2 (EC 2.8.2.-) (HS6ST-2). | |||||
|
LATS1_MOUSE
|
||||||
| NC score | 0.008281 (rank : 66) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
SIP1_MOUSE
|
||||||
| NC score | 0.005354 (rank : 67) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0G7 | Gene names | Zfhx1b, Sip1, Zfx1b | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 1b (Smad-interacting protein 1). | |||||
|
SIP1_HUMAN
|
||||||
| NC score | 0.005241 (rank : 68) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60315, Q9UED1 | Gene names | ZFHX1B, KIAA0569, SIP1, ZFX1B | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 1b (Smad-interacting protein 1) (SMADIP1). | |||||
|
SN1L2_HUMAN
|
||||||
| NC score | 0.004043 (rank : 69) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0K1, O94878, Q6AZE2, Q76N03, Q8NCV7, Q96CZ8 | Gene names | SNF1LK2, KIAA0781, QIK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 2 (EC 2.7.11.1) (Qin- induced kinase). | |||||
|
PCD15_MOUSE
|
||||||
| NC score | 0.003858 (rank : 70) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99PJ1 | Gene names | Pcdh15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
MAK_MOUSE
|
||||||
| NC score | 0.000842 (rank : 71) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04859 | Gene names | Mak, Rck | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MAK (EC 2.7.11.22) (Male germ cell- associated kinase) (Protein kinase RCK). | |||||