Please be patient as the page loads
|
PIGQ_MOUSE
|
||||||
| SwissProt Accessions | Q9QYT7, O35120, O35456, Q99L11 | Gene names | Pigq, Gpi1h, Mgpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q (EC 2.4.1.198) (Phosphatidylinositol-glycan biosynthesis class Q protein) (PIG-Q) (N-acetylglucosamyl transferase component GPI1) (MGpi1p). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PIGQ_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.965759 (rank : 2) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BRB3, O14927, Q96G00, Q96S22, Q9UJH4 | Gene names | PIGQ, GPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q (EC 2.4.1.198) (Phosphatidylinositol-glycan biosynthesis class Q protein) (PIG-Q) (N-acetylglucosamyl transferase component GPI1). | |||||
|
PIGQ_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QYT7, O35120, O35456, Q99L11 | Gene names | Pigq, Gpi1h, Mgpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q (EC 2.4.1.198) (Phosphatidylinositol-glycan biosynthesis class Q protein) (PIG-Q) (N-acetylglucosamyl transferase component GPI1) (MGpi1p). | |||||
|
KRIT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 3) | NC score | 0.012719 (rank : 4) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00522, O43894, Q506L6, Q6U276, Q75N19, Q9H180, Q9H264, Q9HAX5 | Gene names | KRIT1, CCM1 | |||
|
Domain Architecture |
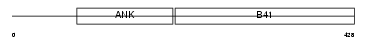
|
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein). | |||||
|
ENT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 4) | NC score | 0.019231 (rank : 3) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P61550 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-T_19q13.11 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-T Env protein) [Includes: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
LCAP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 5) | NC score | 0.009817 (rank : 5) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UIQ6, O00769, Q15145, Q9UIQ7 | Gene names | LNPEP, OTASE | |||
|
Domain Architecture |
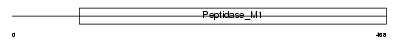
|
|||||
| Description | Leucyl-cystinyl aminopeptidase (EC 3.4.11.3) (Cystinyl aminopeptidase) (Oxytocinase) (OTase) (Insulin-regulated membrane aminopeptidase) (Insulin-responsive aminopeptidase) (IRAP) (Placental leucine aminopeptidase) (P-LAP). | |||||
|
PIGQ_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QYT7, O35120, O35456, Q99L11 | Gene names | Pigq, Gpi1h, Mgpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q (EC 2.4.1.198) (Phosphatidylinositol-glycan biosynthesis class Q protein) (PIG-Q) (N-acetylglucosamyl transferase component GPI1) (MGpi1p). | |||||
|
PIGQ_HUMAN
|
||||||
| NC score | 0.965759 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BRB3, O14927, Q96G00, Q96S22, Q9UJH4 | Gene names | PIGQ, GPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q (EC 2.4.1.198) (Phosphatidylinositol-glycan biosynthesis class Q protein) (PIG-Q) (N-acetylglucosamyl transferase component GPI1). | |||||
|
ENT1_HUMAN
|
||||||
| NC score | 0.019231 (rank : 3) | θ value | 6.88961 (rank : 4) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P61550 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-T_19q13.11 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-T Env protein) [Includes: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
KRIT1_HUMAN
|
||||||
| NC score | 0.012719 (rank : 4) | θ value | 5.27518 (rank : 3) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00522, O43894, Q506L6, Q6U276, Q75N19, Q9H180, Q9H264, Q9HAX5 | Gene names | KRIT1, CCM1 | |||
|
Domain Architecture |

|
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein). | |||||
|
LCAP_HUMAN
|
||||||
| NC score | 0.009817 (rank : 5) | θ value | 6.88961 (rank : 5) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UIQ6, O00769, Q15145, Q9UIQ7 | Gene names | LNPEP, OTASE | |||
|
Domain Architecture |

|
|||||
| Description | Leucyl-cystinyl aminopeptidase (EC 3.4.11.3) (Cystinyl aminopeptidase) (Oxytocinase) (OTase) (Insulin-regulated membrane aminopeptidase) (Insulin-responsive aminopeptidase) (IRAP) (Placental leucine aminopeptidase) (P-LAP). | |||||