Please be patient as the page loads
|
PEPA_HUMAN
|
||||||
| SwissProt Accessions | P00790, Q8N1E3 | Gene names | PGA3 | |||
|
Domain Architecture |
|
|||||
| Description | Pepsin A precursor (EC 3.4.23.1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PEPA_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P00790, Q8N1E3 | Gene names | PGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Pepsin A precursor (EC 3.4.23.1). | |||||
|
PEPC_MOUSE
|
||||||
| θ value | 3.91003e-112 (rank : 2) | NC score | 0.991896 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D7R7, Q9D7T2 | Gene names | Pgc, Upg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gastricsin precursor (EC 3.4.23.3) (Pepsinogen C). | |||||
|
CATE_HUMAN
|
||||||
| θ value | 6.6695e-112 (rank : 3) | NC score | 0.992191 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P14091, Q9NY58 | Gene names | CTSE | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin E precursor (EC 3.4.23.34). | |||||
|
CATE_MOUSE
|
||||||
| θ value | 1.64275e-110 (rank : 4) | NC score | 0.991776 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70269, O35647 | Gene names | Ctse | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin E precursor (EC 3.4.23.34). | |||||
|
PEPC_HUMAN
|
||||||
| θ value | 5.84277e-108 (rank : 5) | NC score | 0.990883 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P20142, Q5T3D7, Q5T3D8 | Gene names | PGC | |||
|
Domain Architecture |
|
|||||
| Description | Gastricsin precursor (EC 3.4.23.3) (Pepsinogen C). | |||||
|
CATD_MOUSE
|
||||||
| θ value | 4.49445e-92 (rank : 6) | NC score | 0.985886 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P18242 | Gene names | Ctsd | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin D precursor (EC 3.4.23.5). | |||||
|
CATD_HUMAN
|
||||||
| θ value | 4.96919e-91 (rank : 7) | NC score | 0.985298 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P07339, Q6IB57 | Gene names | CTSD | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin D precursor (EC 3.4.23.5) [Contains: Cathepsin D light chain; Cathepsin D heavy chain]. | |||||
|
NAPSA_HUMAN
|
||||||
| θ value | 4.08394e-77 (rank : 8) | NC score | 0.976601 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O96009 | Gene names | NAPSA, NAP1 | |||
|
Domain Architecture |
|
|||||
| Description | Napsin-A precursor (EC 3.4.23.-) (Napsin-1) (NAPA) (TA01/TA02) (Aspartyl protease 4) (Asp 4) (ASP4). | |||||
|
NAPSA_MOUSE
|
||||||
| θ value | 9.0981e-77 (rank : 9) | NC score | 0.978685 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O09043 | Gene names | Napsa, Kdap, Nap | |||
|
Domain Architecture |
|
|||||
| Description | Napsin-A precursor (EC 3.4.23.-) (Kidney-derived aspartic protease- like protein) (KDAP-1) (KAP). | |||||
|
RENI2_MOUSE
|
||||||
| θ value | 1.04096e-72 (rank : 10) | NC score | 0.971571 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00796, P70229, P97955, Q62155 | Gene names | Ren2, Ren-2 | |||
|
Domain Architecture |
|
|||||
| Description | Renin-2 precursor (EC 3.4.23.15) (Angiotensinogenase) (Submandibular gland renin) [Contains: Renin-2 heavy chain; Renin-2 light chain]. | |||||
|
RENI1_MOUSE
|
||||||
| θ value | 3.95564e-72 (rank : 11) | NC score | 0.971190 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P06281, P97911, Q543E5, Q62153, Q62154 | Gene names | Ren1, Ren, Ren-1 | |||
|
Domain Architecture |
|
|||||
| Description | Renin-1 precursor (EC 3.4.23.15) (Angiotensinogenase) (Kidney renin). | |||||
|
RENI_HUMAN
|
||||||
| θ value | 6.98233e-69 (rank : 12) | NC score | 0.971552 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00797, Q6T5C2 | Gene names | REN | |||
|
Domain Architecture |
|
|||||
| Description | Renin precursor (EC 3.4.23.15) (Angiotensinogenase). | |||||
|
BACE2_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 13) | NC score | 0.804578 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y5Z0, Q9UJT6 | Gene names | BACE2, ASP21 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-secretase 2 precursor (EC 3.4.23.45) (Beta-site APP-cleaving enzyme 2) (Aspartyl protease 1) (Asp 1) (ASP1) (Membrane-associated aspartic protease 1) (Memapsin-1) (Down region aspartic protease). | |||||
|
BACE1_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 14) | NC score | 0.739401 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56817, Q9BYB9, Q9BYC0, Q9BYC1, Q9UJT5 | Gene names | BACE1, BACE | |||
|
Domain Architecture |
|
|||||
| Description | Beta-secretase 1 precursor (EC 3.4.23.46) (Beta-site APP cleaving enzyme 1) (Beta-site amyloid precursor protein cleaving enzyme 1) (Membrane-associated aspartic protease 2) (Memapsin-2) (Aspartyl protease 2) (Asp 2) (ASP2). | |||||
|
BACE1_MOUSE
|
||||||
| θ value | 7.82807e-20 (rank : 15) | NC score | 0.740406 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56818, Q544D0 | Gene names | Bace1, Bace | |||
|
Domain Architecture |
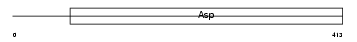
|
|||||
| Description | Beta-secretase 1 precursor (EC 3.4.23.46) (Beta-site APP cleaving enzyme 1) (Beta-site amyloid precursor protein cleaving enzyme 1) (Membrane-associated aspartic protease 2) (Memapsin-2) (Aspartyl protease 2) (Asp 2) (ASP2). | |||||
|
BRCA2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.020626 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
ACPH_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.018839 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13798, Q9BQ33, Q9P0Y2 | Gene names | APEH | |||
|
Domain Architecture |

|
|||||
| Description | Acylamino-acid-releasing enzyme (EC 3.4.19.1) (AARE) (Acyl-peptide hydrolase) (APH) (Acylaminoacyl-peptidase) (Oxidized protein hydrolase) (OPH) (DNF15S2 protein). | |||||
|
NUAK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | -0.002230 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H093 | Gene names | NUAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 2 (EC 2.7.11.1) (SNF1/AMP kinase-related kinase) (SNARK). | |||||
|
TSSK4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | -0.001995 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D411 | Gene names | Tssk4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific serine/threonine-protein kinase 4 (EC 2.7.11.1) (TSSK- 4) (Testis-specific kinase 4) (TSK-4). | |||||
|
LHX4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.001831 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q969G2, Q8NHE0, Q8TCJ1, Q8WWX2, Q969W2 | Gene names | LHX4 | |||
|
Domain Architecture |
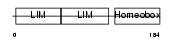
|
|||||
| Description | LIM/homeobox protein Lhx4. | |||||
|
LHX4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.001832 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53776, O08916, Q9R280 | Gene names | Lhx4, Gsh-4, Gsh4 | |||
|
Domain Architecture |
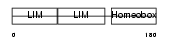
|
|||||
| Description | LIM/homeobox protein Lhx4. | |||||
|
TIF1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.002317 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
PEPA_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P00790, Q8N1E3 | Gene names | PGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Pepsin A precursor (EC 3.4.23.1). | |||||
|
CATE_HUMAN
|
||||||
| NC score | 0.992191 (rank : 2) | θ value | 6.6695e-112 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P14091, Q9NY58 | Gene names | CTSE | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin E precursor (EC 3.4.23.34). | |||||
|
PEPC_MOUSE
|
||||||
| NC score | 0.991896 (rank : 3) | θ value | 3.91003e-112 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D7R7, Q9D7T2 | Gene names | Pgc, Upg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gastricsin precursor (EC 3.4.23.3) (Pepsinogen C). | |||||
|
CATE_MOUSE
|
||||||
| NC score | 0.991776 (rank : 4) | θ value | 1.64275e-110 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70269, O35647 | Gene names | Ctse | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin E precursor (EC 3.4.23.34). | |||||
|
PEPC_HUMAN
|
||||||
| NC score | 0.990883 (rank : 5) | θ value | 5.84277e-108 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P20142, Q5T3D7, Q5T3D8 | Gene names | PGC | |||
|
Domain Architecture |

|
|||||
| Description | Gastricsin precursor (EC 3.4.23.3) (Pepsinogen C). | |||||
|
CATD_MOUSE
|
||||||
| NC score | 0.985886 (rank : 6) | θ value | 4.49445e-92 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P18242 | Gene names | Ctsd | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin D precursor (EC 3.4.23.5). | |||||
|
CATD_HUMAN
|
||||||
| NC score | 0.985298 (rank : 7) | θ value | 4.96919e-91 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P07339, Q6IB57 | Gene names | CTSD | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin D precursor (EC 3.4.23.5) [Contains: Cathepsin D light chain; Cathepsin D heavy chain]. | |||||
|
NAPSA_MOUSE
|
||||||
| NC score | 0.978685 (rank : 8) | θ value | 9.0981e-77 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O09043 | Gene names | Napsa, Kdap, Nap | |||
|
Domain Architecture |

|
|||||
| Description | Napsin-A precursor (EC 3.4.23.-) (Kidney-derived aspartic protease- like protein) (KDAP-1) (KAP). | |||||
|
NAPSA_HUMAN
|
||||||
| NC score | 0.976601 (rank : 9) | θ value | 4.08394e-77 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O96009 | Gene names | NAPSA, NAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Napsin-A precursor (EC 3.4.23.-) (Napsin-1) (NAPA) (TA01/TA02) (Aspartyl protease 4) (Asp 4) (ASP4). | |||||
|
RENI2_MOUSE
|
||||||
| NC score | 0.971571 (rank : 10) | θ value | 1.04096e-72 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00796, P70229, P97955, Q62155 | Gene names | Ren2, Ren-2 | |||
|
Domain Architecture |

|
|||||
| Description | Renin-2 precursor (EC 3.4.23.15) (Angiotensinogenase) (Submandibular gland renin) [Contains: Renin-2 heavy chain; Renin-2 light chain]. | |||||
|
RENI_HUMAN
|
||||||
| NC score | 0.971552 (rank : 11) | θ value | 6.98233e-69 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00797, Q6T5C2 | Gene names | REN | |||
|
Domain Architecture |

|
|||||
| Description | Renin precursor (EC 3.4.23.15) (Angiotensinogenase). | |||||
|
RENI1_MOUSE
|
||||||
| NC score | 0.971190 (rank : 12) | θ value | 3.95564e-72 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P06281, P97911, Q543E5, Q62153, Q62154 | Gene names | Ren1, Ren, Ren-1 | |||
|
Domain Architecture |

|
|||||
| Description | Renin-1 precursor (EC 3.4.23.15) (Angiotensinogenase) (Kidney renin). | |||||
|
BACE2_HUMAN
|
||||||
| NC score | 0.804578 (rank : 13) | θ value | 4.15078e-21 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y5Z0, Q9UJT6 | Gene names | BACE2, ASP21 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-secretase 2 precursor (EC 3.4.23.45) (Beta-site APP-cleaving enzyme 2) (Aspartyl protease 1) (Asp 1) (ASP1) (Membrane-associated aspartic protease 1) (Memapsin-1) (Down region aspartic protease). | |||||
|
BACE1_MOUSE
|
||||||
| NC score | 0.740406 (rank : 14) | θ value | 7.82807e-20 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56818, Q544D0 | Gene names | Bace1, Bace | |||
|
Domain Architecture |

|
|||||
| Description | Beta-secretase 1 precursor (EC 3.4.23.46) (Beta-site APP cleaving enzyme 1) (Beta-site amyloid precursor protein cleaving enzyme 1) (Membrane-associated aspartic protease 2) (Memapsin-2) (Aspartyl protease 2) (Asp 2) (ASP2). | |||||
|
BACE1_HUMAN
|
||||||
| NC score | 0.739401 (rank : 15) | θ value | 3.51386e-20 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56817, Q9BYB9, Q9BYC0, Q9BYC1, Q9UJT5 | Gene names | BACE1, BACE | |||
|
Domain Architecture |

|
|||||
| Description | Beta-secretase 1 precursor (EC 3.4.23.46) (Beta-site APP cleaving enzyme 1) (Beta-site amyloid precursor protein cleaving enzyme 1) (Membrane-associated aspartic protease 2) (Memapsin-2) (Aspartyl protease 2) (Asp 2) (ASP2). | |||||
|
BRCA2_HUMAN
|
||||||
| NC score | 0.020626 (rank : 16) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
ACPH_HUMAN
|
||||||
| NC score | 0.018839 (rank : 17) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13798, Q9BQ33, Q9P0Y2 | Gene names | APEH | |||
|
Domain Architecture |

|
|||||
| Description | Acylamino-acid-releasing enzyme (EC 3.4.19.1) (AARE) (Acyl-peptide hydrolase) (APH) (Acylaminoacyl-peptidase) (Oxidized protein hydrolase) (OPH) (DNF15S2 protein). | |||||
|
TIF1A_MOUSE
|
||||||
| NC score | 0.002317 (rank : 18) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
LHX4_MOUSE
|
||||||
| NC score | 0.001832 (rank : 19) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53776, O08916, Q9R280 | Gene names | Lhx4, Gsh-4, Gsh4 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx4. | |||||
|
LHX4_HUMAN
|
||||||
| NC score | 0.001831 (rank : 20) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q969G2, Q8NHE0, Q8TCJ1, Q8WWX2, Q969W2 | Gene names | LHX4 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx4. | |||||
|
TSSK4_MOUSE
|
||||||
| NC score | -0.001995 (rank : 21) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D411 | Gene names | Tssk4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific serine/threonine-protein kinase 4 (EC 2.7.11.1) (TSSK- 4) (Testis-specific kinase 4) (TSK-4). | |||||
|
NUAK2_HUMAN
|
||||||
| NC score | -0.002230 (rank : 22) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H093 | Gene names | NUAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 2 (EC 2.7.11.1) (SNF1/AMP kinase-related kinase) (SNARK). | |||||