Please be patient as the page loads
|
OC90_HUMAN
|
||||||
| SwissProt Accessions | Q02509 | Gene names | OC90, PLA2L | |||
|
Domain Architecture |

|
|||||
| Description | Otoconin 90 precursor (Oc90) (Phospholipase A2 homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
OC90_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02509 | Gene names | OC90, PLA2L | |||
|
Domain Architecture |

|
|||||
| Description | Otoconin 90 precursor (Oc90) (Phospholipase A2 homolog). | |||||
|
OC90_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.984662 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z0L3, Q9CZ60, Q9Z225 | Gene names | Oc90, Onc-95, Pla2ll | |||
|
Domain Architecture |

|
|||||
| Description | Otoconin 90 precursor (Oc90) (Otoconin-95) (Oc95). | |||||
|
PA2GX_MOUSE
|
||||||
| θ value | 2.97466e-19 (rank : 3) | NC score | 0.795532 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QXX3, Q9EQK6 | Gene names | Pla2g10 | |||
|
Domain Architecture |

|
|||||
| Description | Group 10 secretory phospholipase A2 precursor (EC 3.1.1.4) (Group X secretory phospholipase A2) (Phosphatidylcholine 2-acylhydrolase GX) (GX sPLA2) (sPLA2-X). | |||||
|
PA2GX_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 4) | NC score | 0.786516 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15496, Q6NT23 | Gene names | PLA2G10 | |||
|
Domain Architecture |

|
|||||
| Description | Group 10 secretory phospholipase A2 precursor (EC 3.1.1.4) (Group X secretory phospholipase A2) (Phosphatidylcholine 2-acylhydrolase GX) (GX sPLA2) (sPLA2-X). | |||||
|
PA2GA_MOUSE
|
||||||
| θ value | 1.52774e-15 (rank : 5) | NC score | 0.760648 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P31482, Q60871 | Gene names | Pla2g2a | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2, membrane associated precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (Group IIA phospholipase A2) (GIIC sPLA2) (Enhancing factor) (EF). | |||||
|
PA2GA_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 6) | NC score | 0.754214 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P14555, Q6DN24, Q6IBD9, Q9UCD2 | Gene names | PLA2G2A, PLA2B, PLA2L, RASF-A | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2, membrane associated precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (Group IIA phospholipase A2) (GIIC sPLA2) (Non-pancreatic secretory phospholipase A2) (NPS-PLA2). | |||||
|
PA21B_MOUSE
|
||||||
| θ value | 1.2105e-12 (rank : 7) | NC score | 0.766415 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z0Y2, Q9D7E2, Q9D884 | Gene names | Pla2g1b, Pla2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase) (Group IB phospholipase A2) (PLA2-Ib). | |||||
|
PA2GE_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 8) | NC score | 0.741363 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZK7 | Gene names | PLA2G2E | |||
|
Domain Architecture |

|
|||||
| Description | Group IIE secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIE) (GIIE sPLA2) (sPLA(2)-IIE). | |||||
|
PA21B_HUMAN
|
||||||
| θ value | 2.69671e-12 (rank : 9) | NC score | 0.761976 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04054, Q3KPI1 | Gene names | PLA2G1B, PLA2, PLA2A | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase) (Group IB phospholipase A2). | |||||
|
PA2GD_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 10) | NC score | 0.738015 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVF6, Q3V4B8, Q9JLK0 | Gene names | Pla2g2d, Pla2a2, Splash | |||
|
Domain Architecture |

|
|||||
| Description | Group IID secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIID) (GIID sPLA2) (PLA2IID) (sPLA(2)-IID) (Secretory-type PLA, stroma-associated homolog). | |||||
|
PA2GD_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 11) | NC score | 0.736549 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UNK4, Q9UK01 | Gene names | PLA2G2D, SPLASH | |||
|
Domain Architecture |

|
|||||
| Description | Group IID secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIID) (GIID sPLA2) (PLA2IID) (sPLA(2)-IID) (Secretory-type PLA, stroma-associated homolog). | |||||
|
PA2GE_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 12) | NC score | 0.735830 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QUL3 | Gene names | Pla2g2e | |||
|
Domain Architecture |

|
|||||
| Description | Group IIE secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIE) (GIIE sPLA2) (sPLA(2)-IIE). | |||||
|
PA2GF_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 13) | NC score | 0.736525 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
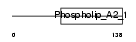
| SwissProt Accessions | Q9BZM2, Q5R385, Q8N217, Q9H506 | Gene names | PLA2G2F | |||
|
Domain Architecture |
|
|||||
| Description | Group IIF secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIF) (GIIF sPLA2) (sPLA(2)-IIF). | |||||
|
PA2GF_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 14) | NC score | 0.734646 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZT4 | Gene names | Pla2g2f | |||
|
Domain Architecture |

|
|||||
| Description | Group IIF secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIF) (GIIF sPLA2) (sPLA(2)-IIF). | |||||
|
PA2GC_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 15) | NC score | 0.732572 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P48076 | Gene names | Pla2g2c | |||
|
Domain Architecture |

|
|||||
| Description | Group IIC secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIC) (GIIC sPLA2) (PLA2-8) (14 kDa phospholipase A2). | |||||
|
PA2G5_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 16) | NC score | 0.697627 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P39877, Q8N435 | Gene names | PLA2G5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-dependent phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (PLA2-10) (Group V phospholipase A2). | |||||
|
PA2G5_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 17) | NC score | 0.705666 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97391, Q9QZU6 | Gene names | Pla2g5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-dependent phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (PLA2-10) (Group V phospholipase A2). | |||||
|
SLIT1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.026835 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80TR4, Q9WVB5 | Gene names | Slit1, Kiaa0813 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 1 protein precursor (Slit-1). | |||||
|
FBN2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.042204 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FBN2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.042209 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FBN3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.038635 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
SLIT1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.027175 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75093, Q8WWZ2, Q9UIL7 | Gene names | SLIT1, KIAA0813, MEGF4, SLIL1 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 1 protein precursor (Slit-1) (Multiple epidermal growth factor-like domains 4). | |||||
|
FA9_HUMAN
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.017163 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 15 | |
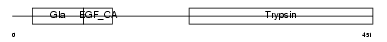
| SwissProt Accessions | P00740 | Gene names | F9 | |||
|
Domain Architecture |
|
|||||
| Description | Coagulation factor IX precursor (EC 3.4.21.22) (Christmas factor) (Plasma thromboplastin component) (PTC) [Contains: Coagulation factor IXa light chain; Coagulation factor IXa heavy chain]. | |||||
|
FBLN2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.032080 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
ADA33_MOUSE
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.014453 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q923W9 | Gene names | Adam33 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 33 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 33). | |||||
|
AHTF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.023138 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CJF7, Q8BVJ5, Q8VD55 | Gene names | Ahctf1, Elys | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
NOTC4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.025256 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
STAB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.025847 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
CUBN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.024659 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
ING3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.015120 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXR8, O60394, Q6GMT3, Q7Z762, Q969G0, Q96DT4, Q9HC99, Q9P081 | Gene names | ING3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
NOTC3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.029264 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
IPPK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.019118 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8X2, Q5T9F7, Q9H7V8 | Gene names | IPPK, C9orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-pentakisphosphate 2-kinase (EC 2.7.1.-) (Inositol-1,3,4,5,6- pentakisphosphate 2-kinase) (Ins(1,3,4,5,6)P5 2-kinase) (InsP5 2- kinase) (IPK1 homolog). | |||||
|
FBLN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.029652 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
SYNG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.013226 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
WFDC5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.021071 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
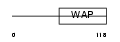
| SwissProt Accessions | Q8TCV5, Q6UWE4 | Gene names | WFDC5, WAP1 | |||
|
Domain Architecture |
|
|||||
| Description | WAP four-disulfide core domain protein 5 precursor (Putative protease inhibitor WAP1). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.023155 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
KR108_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.020719 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
OC90_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02509 | Gene names | OC90, PLA2L | |||
|
Domain Architecture |

|
|||||
| Description | Otoconin 90 precursor (Oc90) (Phospholipase A2 homolog). | |||||
|
OC90_MOUSE
|
||||||
| NC score | 0.984662 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z0L3, Q9CZ60, Q9Z225 | Gene names | Oc90, Onc-95, Pla2ll | |||
|
Domain Architecture |

|
|||||
| Description | Otoconin 90 precursor (Oc90) (Otoconin-95) (Oc95). | |||||
|
PA2GX_MOUSE
|
||||||
| NC score | 0.795532 (rank : 3) | θ value | 2.97466e-19 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QXX3, Q9EQK6 | Gene names | Pla2g10 | |||
|
Domain Architecture |

|
|||||
| Description | Group 10 secretory phospholipase A2 precursor (EC 3.1.1.4) (Group X secretory phospholipase A2) (Phosphatidylcholine 2-acylhydrolase GX) (GX sPLA2) (sPLA2-X). | |||||
|
PA2GX_HUMAN
|
||||||
| NC score | 0.786516 (rank : 4) | θ value | 3.63628e-17 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15496, Q6NT23 | Gene names | PLA2G10 | |||
|
Domain Architecture |

|
|||||
| Description | Group 10 secretory phospholipase A2 precursor (EC 3.1.1.4) (Group X secretory phospholipase A2) (Phosphatidylcholine 2-acylhydrolase GX) (GX sPLA2) (sPLA2-X). | |||||
|
PA21B_MOUSE
|
||||||
| NC score | 0.766415 (rank : 5) | θ value | 1.2105e-12 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z0Y2, Q9D7E2, Q9D884 | Gene names | Pla2g1b, Pla2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase) (Group IB phospholipase A2) (PLA2-Ib). | |||||
|
PA21B_HUMAN
|
||||||
| NC score | 0.761976 (rank : 6) | θ value | 2.69671e-12 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04054, Q3KPI1 | Gene names | PLA2G1B, PLA2, PLA2A | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase) (Group IB phospholipase A2). | |||||
|
PA2GA_MOUSE
|
||||||
| NC score | 0.760648 (rank : 7) | θ value | 1.52774e-15 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P31482, Q60871 | Gene names | Pla2g2a | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2, membrane associated precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (Group IIA phospholipase A2) (GIIC sPLA2) (Enhancing factor) (EF). | |||||
|
PA2GA_HUMAN
|
||||||
| NC score | 0.754214 (rank : 8) | θ value | 3.18553e-13 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P14555, Q6DN24, Q6IBD9, Q9UCD2 | Gene names | PLA2G2A, PLA2B, PLA2L, RASF-A | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2, membrane associated precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (Group IIA phospholipase A2) (GIIC sPLA2) (Non-pancreatic secretory phospholipase A2) (NPS-PLA2). | |||||
|
PA2GE_HUMAN
|
||||||
| NC score | 0.741363 (rank : 9) | θ value | 1.2105e-12 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZK7 | Gene names | PLA2G2E | |||
|
Domain Architecture |

|
|||||
| Description | Group IIE secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIE) (GIIE sPLA2) (sPLA(2)-IIE). | |||||
|
PA2GD_MOUSE
|
||||||
| NC score | 0.738015 (rank : 10) | θ value | 2.69671e-12 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVF6, Q3V4B8, Q9JLK0 | Gene names | Pla2g2d, Pla2a2, Splash | |||
|
Domain Architecture |

|
|||||
| Description | Group IID secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIID) (GIID sPLA2) (PLA2IID) (sPLA(2)-IID) (Secretory-type PLA, stroma-associated homolog). | |||||
|
PA2GD_HUMAN
|
||||||
| NC score | 0.736549 (rank : 11) | θ value | 4.59992e-12 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UNK4, Q9UK01 | Gene names | PLA2G2D, SPLASH | |||
|
Domain Architecture |

|
|||||
| Description | Group IID secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIID) (GIID sPLA2) (PLA2IID) (sPLA(2)-IID) (Secretory-type PLA, stroma-associated homolog). | |||||
|
PA2GF_HUMAN
|
||||||
| NC score | 0.736525 (rank : 12) | θ value | 1.33837e-11 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BZM2, Q5R385, Q8N217, Q9H506 | Gene names | PLA2G2F | |||
|
Domain Architecture |

|
|||||
| Description | Group IIF secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIF) (GIIF sPLA2) (sPLA(2)-IIF). | |||||
|
PA2GE_MOUSE
|
||||||
| NC score | 0.735830 (rank : 13) | θ value | 1.02475e-11 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QUL3 | Gene names | Pla2g2e | |||
|
Domain Architecture |

|
|||||
| Description | Group IIE secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIE) (GIIE sPLA2) (sPLA(2)-IIE). | |||||
|
PA2GF_MOUSE
|
||||||
| NC score | 0.734646 (rank : 14) | θ value | 1.74796e-11 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZT4 | Gene names | Pla2g2f | |||
|
Domain Architecture |

|
|||||
| Description | Group IIF secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIF) (GIIF sPLA2) (sPLA(2)-IIF). | |||||
|
PA2GC_MOUSE
|
||||||
| NC score | 0.732572 (rank : 15) | θ value | 1.25267e-09 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P48076 | Gene names | Pla2g2c | |||
|
Domain Architecture |

|
|||||
| Description | Group IIC secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIC) (GIIC sPLA2) (PLA2-8) (14 kDa phospholipase A2). | |||||
|
PA2G5_MOUSE
|
||||||
| NC score | 0.705666 (rank : 16) | θ value | 4.0297e-08 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97391, Q9QZU6 | Gene names | Pla2g5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-dependent phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (PLA2-10) (Group V phospholipase A2). | |||||
|
PA2G5_HUMAN
|
||||||
| NC score | 0.697627 (rank : 17) | θ value | 1.06045e-08 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P39877, Q8N435 | Gene names | PLA2G5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-dependent phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (PLA2-10) (Group V phospholipase A2). | |||||
|
FBN2_MOUSE
|
||||||
| NC score | 0.042209 (rank : 18) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FBN2_HUMAN
|
||||||
| NC score | 0.042204 (rank : 19) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FBN3_HUMAN
|
||||||
| NC score | 0.038635 (rank : 20) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
FBLN2_MOUSE
|
||||||
| NC score | 0.032080 (rank : 21) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
FBLN2_HUMAN
|
||||||
| NC score | 0.029652 (rank : 22) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
NOTC3_MOUSE
|
||||||
| NC score | 0.029264 (rank : 23) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
SLIT1_HUMAN
|
||||||
| NC score | 0.027175 (rank : 24) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75093, Q8WWZ2, Q9UIL7 | Gene names | SLIT1, KIAA0813, MEGF4, SLIL1 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 1 protein precursor (Slit-1) (Multiple epidermal growth factor-like domains 4). | |||||
|
SLIT1_MOUSE
|
||||||
| NC score | 0.026835 (rank : 25) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80TR4, Q9WVB5 | Gene names | Slit1, Kiaa0813 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 1 protein precursor (Slit-1). | |||||
|
STAB2_MOUSE
|
||||||
| NC score | 0.025847 (rank : 26) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
NOTC4_MOUSE
|
||||||
| NC score | 0.025256 (rank : 27) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
CUBN_MOUSE
|
||||||
| NC score | 0.024659 (rank : 28) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.023155 (rank : 29) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
AHTF1_MOUSE
|
||||||
| NC score | 0.023138 (rank : 30) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CJF7, Q8BVJ5, Q8VD55 | Gene names | Ahctf1, Elys | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
WFDC5_HUMAN
|
||||||
| NC score | 0.021071 (rank : 31) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TCV5, Q6UWE4 | Gene names | WFDC5, WAP1 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 5 precursor (Putative protease inhibitor WAP1). | |||||
|
KR108_HUMAN
|
||||||
| NC score | 0.020719 (rank : 32) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
IPPK_HUMAN
|
||||||
| NC score | 0.019118 (rank : 33) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8X2, Q5T9F7, Q9H7V8 | Gene names | IPPK, C9orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-pentakisphosphate 2-kinase (EC 2.7.1.-) (Inositol-1,3,4,5,6- pentakisphosphate 2-kinase) (Ins(1,3,4,5,6)P5 2-kinase) (InsP5 2- kinase) (IPK1 homolog). | |||||
|
FA9_HUMAN
|
||||||
| NC score | 0.017163 (rank : 34) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00740 | Gene names | F9 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor IX precursor (EC 3.4.21.22) (Christmas factor) (Plasma thromboplastin component) (PTC) [Contains: Coagulation factor IXa light chain; Coagulation factor IXa heavy chain]. | |||||
|
ING3_HUMAN
|
||||||
| NC score | 0.015120 (rank : 35) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXR8, O60394, Q6GMT3, Q7Z762, Q969G0, Q96DT4, Q9HC99, Q9P081 | Gene names | ING3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
ADA33_MOUSE
|
||||||
| NC score | 0.014453 (rank : 36) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q923W9 | Gene names | Adam33 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 33 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 33). | |||||
|
SYNG_MOUSE
|
||||||
| NC score | 0.013226 (rank : 37) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||