Please be patient as the page loads
|
PA2GX_HUMAN
|
||||||
| SwissProt Accessions | O15496, Q6NT23 | Gene names | PLA2G10 | |||
|
Domain Architecture |

|
|||||
| Description | Group 10 secretory phospholipase A2 precursor (EC 3.1.1.4) (Group X secretory phospholipase A2) (Phosphatidylcholine 2-acylhydrolase GX) (GX sPLA2) (sPLA2-X). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PA2GX_HUMAN
|
||||||
| θ value | 1.89267e-82 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15496, Q6NT23 | Gene names | PLA2G10 | |||
|
Domain Architecture |

|
|||||
| Description | Group 10 secretory phospholipase A2 precursor (EC 3.1.1.4) (Group X secretory phospholipase A2) (Phosphatidylcholine 2-acylhydrolase GX) (GX sPLA2) (sPLA2-X). | |||||
|
PA2GX_MOUSE
|
||||||
| θ value | 1.04338e-64 (rank : 2) | NC score | 0.994285 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QXX3, Q9EQK6 | Gene names | Pla2g10 | |||
|
Domain Architecture |

|
|||||
| Description | Group 10 secretory phospholipase A2 precursor (EC 3.1.1.4) (Group X secretory phospholipase A2) (Phosphatidylcholine 2-acylhydrolase GX) (GX sPLA2) (sPLA2-X). | |||||
|
PA2GA_MOUSE
|
||||||
| θ value | 3.87602e-27 (rank : 3) | NC score | 0.951113 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P31482, Q60871 | Gene names | Pla2g2a | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2, membrane associated precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (Group IIA phospholipase A2) (GIIC sPLA2) (Enhancing factor) (EF). | |||||
|
PA2GE_HUMAN
|
||||||
| θ value | 3.62785e-25 (rank : 4) | NC score | 0.947524 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZK7 | Gene names | PLA2G2E | |||
|
Domain Architecture |

|
|||||
| Description | Group IIE secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIE) (GIIE sPLA2) (sPLA(2)-IIE). | |||||
|
PA2GE_MOUSE
|
||||||
| θ value | 4.01107e-24 (rank : 5) | NC score | 0.944780 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QUL3 | Gene names | Pla2g2e | |||
|
Domain Architecture |

|
|||||
| Description | Group IIE secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIE) (GIIE sPLA2) (sPLA(2)-IIE). | |||||
|
PA2GF_MOUSE
|
||||||
| θ value | 2.59989e-23 (rank : 6) | NC score | 0.929056 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZT4 | Gene names | Pla2g2f | |||
|
Domain Architecture |

|
|||||
| Description | Group IIF secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIF) (GIIF sPLA2) (sPLA(2)-IIF). | |||||
|
PA2GD_MOUSE
|
||||||
| θ value | 3.39556e-23 (rank : 7) | NC score | 0.941703 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVF6, Q3V4B8, Q9JLK0 | Gene names | Pla2g2d, Pla2a2, Splash | |||
|
Domain Architecture |

|
|||||
| Description | Group IID secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIID) (GIID sPLA2) (PLA2IID) (sPLA(2)-IID) (Secretory-type PLA, stroma-associated homolog). | |||||
|
PA2GF_HUMAN
|
||||||
| θ value | 7.56453e-23 (rank : 8) | NC score | 0.927846 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BZM2, Q5R385, Q8N217, Q9H506 | Gene names | PLA2G2F | |||
|
Domain Architecture |
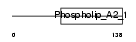
|
|||||
| Description | Group IIF secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIF) (GIIF sPLA2) (sPLA(2)-IIF). | |||||
|
PA2GD_HUMAN
|
||||||
| θ value | 9.87957e-23 (rank : 9) | NC score | 0.940076 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UNK4, Q9UK01 | Gene names | PLA2G2D, SPLASH | |||
|
Domain Architecture |

|
|||||
| Description | Group IID secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIID) (GIID sPLA2) (PLA2IID) (sPLA(2)-IID) (Secretory-type PLA, stroma-associated homolog). | |||||
|
PA2GA_HUMAN
|
||||||
| θ value | 8.36355e-22 (rank : 10) | NC score | 0.944846 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P14555, Q6DN24, Q6IBD9, Q9UCD2 | Gene names | PLA2G2A, PLA2B, PLA2L, RASF-A | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2, membrane associated precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (Group IIA phospholipase A2) (GIIC sPLA2) (Non-pancreatic secretory phospholipase A2) (NPS-PLA2). | |||||
|
PA2G5_MOUSE
|
||||||
| θ value | 9.24701e-21 (rank : 11) | NC score | 0.922108 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97391, Q9QZU6 | Gene names | Pla2g5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-dependent phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (PLA2-10) (Group V phospholipase A2). | |||||
|
PA2G5_HUMAN
|
||||||
| θ value | 2.06002e-20 (rank : 12) | NC score | 0.920485 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P39877, Q8N435 | Gene names | PLA2G5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-dependent phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (PLA2-10) (Group V phospholipase A2). | |||||
|
PA21B_MOUSE
|
||||||
| θ value | 2.27762e-19 (rank : 13) | NC score | 0.903694 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z0Y2, Q9D7E2, Q9D884 | Gene names | Pla2g1b, Pla2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase) (Group IB phospholipase A2) (PLA2-Ib). | |||||
|
PA2GC_MOUSE
|
||||||
| θ value | 4.29542e-18 (rank : 14) | NC score | 0.928969 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P48076 | Gene names | Pla2g2c | |||
|
Domain Architecture |

|
|||||
| Description | Group IIC secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIC) (GIIC sPLA2) (PLA2-8) (14 kDa phospholipase A2). | |||||
|
PA21B_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 15) | NC score | 0.895915 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04054, Q3KPI1 | Gene names | PLA2G1B, PLA2, PLA2A | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase) (Group IB phospholipase A2). | |||||
|
OC90_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 16) | NC score | 0.786516 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q02509 | Gene names | OC90, PLA2L | |||
|
Domain Architecture |

|
|||||
| Description | Otoconin 90 precursor (Oc90) (Phospholipase A2 homolog). | |||||
|
OC90_MOUSE
|
||||||
| θ value | 2.35696e-16 (rank : 17) | NC score | 0.783753 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z0L3, Q9CZ60, Q9Z225 | Gene names | Oc90, Onc-95, Pla2ll | |||
|
Domain Architecture |

|
|||||
| Description | Otoconin 90 precursor (Oc90) (Otoconin-95) (Oc95). | |||||
|
LRP8_MOUSE
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.037555 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q924X6, Q8CAK9, Q8CDF5, Q921B6 | Gene names | Lrp8, Apoer2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
DPF1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.018383 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
LRP8_HUMAN
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.037386 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14114, O14968, Q86V27, Q99876, Q9BR78 | Gene names | LRP8, APOER2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
CORIN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.018161 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y5Q5, Q9UHY2 | Gene names | CORIN, CRN, TMPRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Heart-specific serine proteinase ATC2) (Transmembrane protease, serine 10). | |||||
|
STAB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.021543 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
REQU_MOUSE
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.017515 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
LRP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.032381 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
ADM1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.007482 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60813, Q60617, Q80WR7, Q8R533 | Gene names | Adam1a, Adam1, Ftna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 1a precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 1a) (Fertilin alpha 1a subunit). | |||||
|
CORIN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.017275 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
KRA51_HUMAN
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.023385 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
LDLR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.033377 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P01130, Q53ZD9 | Gene names | LDLR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor precursor (LDL receptor). | |||||
|
LRP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.030037 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
PGBM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.015232 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.022428 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
STAB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.016863 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
GRN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.023079 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
LRP1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.029637 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
PGBM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.013953 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
LRP1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.029680 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
PA2GX_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.89267e-82 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15496, Q6NT23 | Gene names | PLA2G10 | |||
|
Domain Architecture |

|
|||||
| Description | Group 10 secretory phospholipase A2 precursor (EC 3.1.1.4) (Group X secretory phospholipase A2) (Phosphatidylcholine 2-acylhydrolase GX) (GX sPLA2) (sPLA2-X). | |||||
|
PA2GX_MOUSE
|
||||||
| NC score | 0.994285 (rank : 2) | θ value | 1.04338e-64 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QXX3, Q9EQK6 | Gene names | Pla2g10 | |||
|
Domain Architecture |

|
|||||
| Description | Group 10 secretory phospholipase A2 precursor (EC 3.1.1.4) (Group X secretory phospholipase A2) (Phosphatidylcholine 2-acylhydrolase GX) (GX sPLA2) (sPLA2-X). | |||||
|
PA2GA_MOUSE
|
||||||
| NC score | 0.951113 (rank : 3) | θ value | 3.87602e-27 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P31482, Q60871 | Gene names | Pla2g2a | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2, membrane associated precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (Group IIA phospholipase A2) (GIIC sPLA2) (Enhancing factor) (EF). | |||||
|
PA2GE_HUMAN
|
||||||
| NC score | 0.947524 (rank : 4) | θ value | 3.62785e-25 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZK7 | Gene names | PLA2G2E | |||
|
Domain Architecture |

|
|||||
| Description | Group IIE secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIE) (GIIE sPLA2) (sPLA(2)-IIE). | |||||
|
PA2GA_HUMAN
|
||||||
| NC score | 0.944846 (rank : 5) | θ value | 8.36355e-22 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P14555, Q6DN24, Q6IBD9, Q9UCD2 | Gene names | PLA2G2A, PLA2B, PLA2L, RASF-A | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2, membrane associated precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (Group IIA phospholipase A2) (GIIC sPLA2) (Non-pancreatic secretory phospholipase A2) (NPS-PLA2). | |||||
|
PA2GE_MOUSE
|
||||||
| NC score | 0.944780 (rank : 6) | θ value | 4.01107e-24 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QUL3 | Gene names | Pla2g2e | |||
|
Domain Architecture |

|
|||||
| Description | Group IIE secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIE) (GIIE sPLA2) (sPLA(2)-IIE). | |||||
|
PA2GD_MOUSE
|
||||||
| NC score | 0.941703 (rank : 7) | θ value | 3.39556e-23 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVF6, Q3V4B8, Q9JLK0 | Gene names | Pla2g2d, Pla2a2, Splash | |||
|
Domain Architecture |

|
|||||
| Description | Group IID secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIID) (GIID sPLA2) (PLA2IID) (sPLA(2)-IID) (Secretory-type PLA, stroma-associated homolog). | |||||
|
PA2GD_HUMAN
|
||||||
| NC score | 0.940076 (rank : 8) | θ value | 9.87957e-23 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UNK4, Q9UK01 | Gene names | PLA2G2D, SPLASH | |||
|
Domain Architecture |

|
|||||
| Description | Group IID secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIID) (GIID sPLA2) (PLA2IID) (sPLA(2)-IID) (Secretory-type PLA, stroma-associated homolog). | |||||
|
PA2GF_MOUSE
|
||||||
| NC score | 0.929056 (rank : 9) | θ value | 2.59989e-23 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZT4 | Gene names | Pla2g2f | |||
|
Domain Architecture |

|
|||||
| Description | Group IIF secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIF) (GIIF sPLA2) (sPLA(2)-IIF). | |||||
|
PA2GC_MOUSE
|
||||||
| NC score | 0.928969 (rank : 10) | θ value | 4.29542e-18 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P48076 | Gene names | Pla2g2c | |||
|
Domain Architecture |

|
|||||
| Description | Group IIC secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIC) (GIIC sPLA2) (PLA2-8) (14 kDa phospholipase A2). | |||||
|
PA2GF_HUMAN
|
||||||
| NC score | 0.927846 (rank : 11) | θ value | 7.56453e-23 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BZM2, Q5R385, Q8N217, Q9H506 | Gene names | PLA2G2F | |||
|
Domain Architecture |

|
|||||
| Description | Group IIF secretory phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase GIIF) (GIIF sPLA2) (sPLA(2)-IIF). | |||||
|
PA2G5_MOUSE
|
||||||
| NC score | 0.922108 (rank : 12) | θ value | 9.24701e-21 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97391, Q9QZU6 | Gene names | Pla2g5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-dependent phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (PLA2-10) (Group V phospholipase A2). | |||||
|
PA2G5_HUMAN
|
||||||
| NC score | 0.920485 (rank : 13) | θ value | 2.06002e-20 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P39877, Q8N435 | Gene names | PLA2G5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-dependent phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2-acylhydrolase) (PLA2-10) (Group V phospholipase A2). | |||||
|
PA21B_MOUSE
|
||||||
| NC score | 0.903694 (rank : 14) | θ value | 2.27762e-19 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z0Y2, Q9D7E2, Q9D884 | Gene names | Pla2g1b, Pla2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase) (Group IB phospholipase A2) (PLA2-Ib). | |||||
|
PA21B_HUMAN
|
||||||
| NC score | 0.895915 (rank : 15) | θ value | 2.7842e-17 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04054, Q3KPI1 | Gene names | PLA2G1B, PLA2, PLA2A | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase A2 precursor (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase) (Group IB phospholipase A2). | |||||
|
OC90_HUMAN
|
||||||
| NC score | 0.786516 (rank : 16) | θ value | 3.63628e-17 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q02509 | Gene names | OC90, PLA2L | |||
|
Domain Architecture |

|
|||||
| Description | Otoconin 90 precursor (Oc90) (Phospholipase A2 homolog). | |||||
|
OC90_MOUSE
|
||||||
| NC score | 0.783753 (rank : 17) | θ value | 2.35696e-16 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z0L3, Q9CZ60, Q9Z225 | Gene names | Oc90, Onc-95, Pla2ll | |||
|
Domain Architecture |

|
|||||
| Description | Otoconin 90 precursor (Oc90) (Otoconin-95) (Oc95). | |||||
|
LRP8_MOUSE
|
||||||
| NC score | 0.037555 (rank : 18) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q924X6, Q8CAK9, Q8CDF5, Q921B6 | Gene names | Lrp8, Apoer2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
LRP8_HUMAN
|
||||||
| NC score | 0.037386 (rank : 19) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14114, O14968, Q86V27, Q99876, Q9BR78 | Gene names | LRP8, APOER2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
LDLR_HUMAN
|
||||||
| NC score | 0.033377 (rank : 20) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P01130, Q53ZD9 | Gene names | LDLR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor precursor (LDL receptor). | |||||
|
LRP2_HUMAN
|
||||||
| NC score | 0.032381 (rank : 21) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
LRP1_HUMAN
|
||||||
| NC score | 0.030037 (rank : 22) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
LRP1B_MOUSE
|
||||||
| NC score | 0.029680 (rank : 23) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
LRP1B_HUMAN
|
||||||
| NC score | 0.029637 (rank : 24) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
KRA51_HUMAN
|
||||||
| NC score | 0.023385 (rank : 25) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
GRN_HUMAN
|
||||||
| NC score | 0.023079 (rank : 26) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.022428 (rank : 27) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
STAB1_HUMAN
|
||||||
| NC score | 0.021543 (rank : 28) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
DPF1_MOUSE
|
||||||
| NC score | 0.018383 (rank : 29) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
CORIN_HUMAN
|
||||||
| NC score | 0.018161 (rank : 30) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y5Q5, Q9UHY2 | Gene names | CORIN, CRN, TMPRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Heart-specific serine proteinase ATC2) (Transmembrane protease, serine 10). | |||||
|
REQU_MOUSE
|
||||||
| NC score | 0.017515 (rank : 31) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
CORIN_MOUSE
|
||||||
| NC score | 0.017275 (rank : 32) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
STAB1_MOUSE
|
||||||
| NC score | 0.016863 (rank : 33) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
PGBM_MOUSE
|
||||||
| NC score | 0.015232 (rank : 34) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.013953 (rank : 35) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
ADM1A_MOUSE
|
||||||
| NC score | 0.007482 (rank : 36) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60813, Q60617, Q80WR7, Q8R533 | Gene names | Adam1a, Adam1, Ftna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 1a precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 1a) (Fertilin alpha 1a subunit). | |||||