Please be patient as the page loads
|
NNTM_HUMAN
|
||||||
| SwissProt Accessions | Q13423, Q16796, Q8N3V4 | Gene names | NNT | |||
|
Domain Architecture |

|
|||||
| Description | NAD(P) transhydrogenase, mitochondrial precursor (EC 1.6.1.2) (Pyridine nucleotide transhydrogenase) (Nicotinamide nucleotide transhydrogenase). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NNTM_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13423, Q16796, Q8N3V4 | Gene names | NNT | |||
|
Domain Architecture |

|
|||||
| Description | NAD(P) transhydrogenase, mitochondrial precursor (EC 1.6.1.2) (Pyridine nucleotide transhydrogenase) (Nicotinamide nucleotide transhydrogenase). | |||||
|
NNTM_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998796 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61941, Q9JK70 | Gene names | Nnt | |||
|
Domain Architecture |

|
|||||
| Description | NAD(P) transhydrogenase, mitochondrial precursor (EC 1.6.1.2) (Pyridine nucleotide transhydrogenase) (Nicotinamide nucleotide transhydrogenase). | |||||
|
TMM65_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 3) | NC score | 0.107419 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PI78, Q8N5G8, Q8WVK5 | Gene names | TMEM65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 65. | |||||
|
GTR4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 4) | NC score | 0.034535 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14142, Q9JJN9 | Gene names | Slc2a4, Glut-4, Glut4 | |||
|
Domain Architecture |
|
|||||
| Description | Solute carrier family 2, facilitated glucose transporter member 4 (Glucose transporter type 4, insulin-responsive) (GT2). | |||||
|
GTR4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 5) | NC score | 0.032896 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14672 | Gene names | SLC2A4, GLUT4 | |||
|
Domain Architecture |
|
|||||
| Description | Solute carrier family 2, facilitated glucose transporter member 4 (Glucose transporter type 4, insulin-responsive). | |||||
|
S23A2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 6) | NC score | 0.043725 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UGH3, Q8WWR4, Q92512, Q96D54, Q9UNU1, Q9UP85 | Gene names | SLC23A2, KIAA0238, NBTL1, SVCT2, YSPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 23 member 2 (Sodium-dependent vitamin C transporter 2) (hSVCT2) (Na(+)/L-ascorbic acid transporter 2) (Yolk sac permease-like molecule 2) (Nucleobase transporter-like 1 protein). | |||||
|
S23A2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 7) | NC score | 0.041311 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPR4, Q8C327, Q9JM78 | Gene names | Slc23a2, Kiaa0238, Svct2, Yspl2 | |||
|
Domain Architecture |
|
|||||
| Description | Solute carrier family 23 member 2 (Sodium-dependent vitamin C transporter 2) (mSVCT2) (Na(+)/L-ascorbic acid transporter 2) (Yolk sac permease-like molecule 2). | |||||
|
DHSO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 8) | NC score | 0.036622 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00796, Q16682, Q9UMD6 | Gene names | SORD | |||
|
Domain Architecture |
|
|||||
| Description | Sorbitol dehydrogenase (EC 1.1.1.14) (L-iditol 2-dehydrogenase). | |||||
|
ETFD_MOUSE
|
||||||
| θ value | 4.03905 (rank : 9) | NC score | 0.042334 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q921G7, Q3U7K2, Q8BK82, Q9DCT9 | Gene names | Etfdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial precursor (EC 1.5.5.1) (ETF-QO) (ETF-ubiquinone oxidoreductase) (ETF dehydrogenase) (Electron-transferring- flavoprotein dehydrogenase). | |||||
|
BRDT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 10) | NC score | 0.016886 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 5.27518 (rank : 11) | NC score | 0.027442 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
MINT_MOUSE
|
||||||
| θ value | 5.27518 (rank : 12) | NC score | 0.009500 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
OPLA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 13) | NC score | 0.034770 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K010, Q8R3K2 | Gene names | Oplah | |||
|
Domain Architecture |

|
|||||
| Description | 5-oxoprolinase (EC 3.5.2.9) (5-oxo-L-prolinase) (Pyroglutamase) (5- OPase). | |||||
|
SIRT5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 14) | NC score | 0.026520 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXA8, Q9Y6E6 | Gene names | SIRT5, SIR2L5 | |||
|
Domain Architecture |
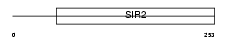
|
|||||
| Description | NAD-dependent deacetylase sirtuin-5 (EC 3.5.1.-) (SIR2-like protein 5). | |||||
|
ETFD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 15) | NC score | 0.033954 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16134, Q7Z347 | Gene names | ETFDH | |||
|
Domain Architecture |

|
|||||
| Description | Electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial precursor (EC 1.5.5.1) (ETF-QO) (ETF-ubiquinone oxidoreductase) (ETF dehydrogenase) (Electron-transferring- flavoprotein dehydrogenase). | |||||
|
JAD1D_HUMAN
|
||||||
| θ value | 8.99809 (rank : 16) | NC score | 0.006399 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
OPLA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.028861 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14841, Q75W65, Q9Y4Q0 | Gene names | OPLAH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-oxoprolinase (EC 3.5.2.9) (5-oxo-L-prolinase) (Pyroglutamase) (5- OPase). | |||||
|
NNTM_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13423, Q16796, Q8N3V4 | Gene names | NNT | |||
|
Domain Architecture |

|
|||||
| Description | NAD(P) transhydrogenase, mitochondrial precursor (EC 1.6.1.2) (Pyridine nucleotide transhydrogenase) (Nicotinamide nucleotide transhydrogenase). | |||||
|
NNTM_MOUSE
|
||||||
| NC score | 0.998796 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61941, Q9JK70 | Gene names | Nnt | |||
|
Domain Architecture |

|
|||||
| Description | NAD(P) transhydrogenase, mitochondrial precursor (EC 1.6.1.2) (Pyridine nucleotide transhydrogenase) (Nicotinamide nucleotide transhydrogenase). | |||||
|
TMM65_HUMAN
|
||||||
| NC score | 0.107419 (rank : 3) | θ value | 0.0736092 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PI78, Q8N5G8, Q8WVK5 | Gene names | TMEM65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 65. | |||||
|
S23A2_HUMAN
|
||||||
| NC score | 0.043725 (rank : 4) | θ value | 2.36792 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UGH3, Q8WWR4, Q92512, Q96D54, Q9UNU1, Q9UP85 | Gene names | SLC23A2, KIAA0238, NBTL1, SVCT2, YSPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 23 member 2 (Sodium-dependent vitamin C transporter 2) (hSVCT2) (Na(+)/L-ascorbic acid transporter 2) (Yolk sac permease-like molecule 2) (Nucleobase transporter-like 1 protein). | |||||
|
ETFD_MOUSE
|
||||||
| NC score | 0.042334 (rank : 5) | θ value | 4.03905 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q921G7, Q3U7K2, Q8BK82, Q9DCT9 | Gene names | Etfdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial precursor (EC 1.5.5.1) (ETF-QO) (ETF-ubiquinone oxidoreductase) (ETF dehydrogenase) (Electron-transferring- flavoprotein dehydrogenase). | |||||
|
S23A2_MOUSE
|
||||||
| NC score | 0.041311 (rank : 6) | θ value | 3.0926 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPR4, Q8C327, Q9JM78 | Gene names | Slc23a2, Kiaa0238, Svct2, Yspl2 | |||
|
Domain Architecture |
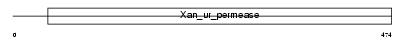
|
|||||
| Description | Solute carrier family 23 member 2 (Sodium-dependent vitamin C transporter 2) (mSVCT2) (Na(+)/L-ascorbic acid transporter 2) (Yolk sac permease-like molecule 2). | |||||
|
DHSO_HUMAN
|
||||||
| NC score | 0.036622 (rank : 7) | θ value | 4.03905 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00796, Q16682, Q9UMD6 | Gene names | SORD | |||
|
Domain Architecture |
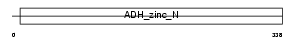
|
|||||
| Description | Sorbitol dehydrogenase (EC 1.1.1.14) (L-iditol 2-dehydrogenase). | |||||
|
OPLA_MOUSE
|
||||||
| NC score | 0.034770 (rank : 8) | θ value | 5.27518 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K010, Q8R3K2 | Gene names | Oplah | |||
|
Domain Architecture |

|
|||||
| Description | 5-oxoprolinase (EC 3.5.2.9) (5-oxo-L-prolinase) (Pyroglutamase) (5- OPase). | |||||
|
GTR4_MOUSE
|
||||||
| NC score | 0.034535 (rank : 9) | θ value | 1.38821 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14142, Q9JJN9 | Gene names | Slc2a4, Glut-4, Glut4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 2, facilitated glucose transporter member 4 (Glucose transporter type 4, insulin-responsive) (GT2). | |||||
|
ETFD_HUMAN
|
||||||
| NC score | 0.033954 (rank : 10) | θ value | 8.99809 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16134, Q7Z347 | Gene names | ETFDH | |||
|
Domain Architecture |

|
|||||
| Description | Electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial precursor (EC 1.5.5.1) (ETF-QO) (ETF-ubiquinone oxidoreductase) (ETF dehydrogenase) (Electron-transferring- flavoprotein dehydrogenase). | |||||
|
GTR4_HUMAN
|
||||||
| NC score | 0.032896 (rank : 11) | θ value | 1.81305 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14672 | Gene names | SLC2A4, GLUT4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 2, facilitated glucose transporter member 4 (Glucose transporter type 4, insulin-responsive). | |||||
|
OPLA_HUMAN
|
||||||
| NC score | 0.028861 (rank : 12) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14841, Q75W65, Q9Y4Q0 | Gene names | OPLAH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5-oxoprolinase (EC 3.5.2.9) (5-oxo-L-prolinase) (Pyroglutamase) (5- OPase). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.027442 (rank : 13) | θ value | 5.27518 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
SIRT5_HUMAN
|
||||||
| NC score | 0.026520 (rank : 14) | θ value | 5.27518 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXA8, Q9Y6E6 | Gene names | SIRT5, SIR2L5 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-5 (EC 3.5.1.-) (SIR2-like protein 5). | |||||
|
BRDT_HUMAN
|
||||||
| NC score | 0.016886 (rank : 15) | θ value | 5.27518 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.009500 (rank : 16) | θ value | 5.27518 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
JAD1D_HUMAN
|
||||||
| NC score | 0.006399 (rank : 17) | θ value | 8.99809 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||