Please be patient as the page loads
|
NBR1_HUMAN
|
||||||
| SwissProt Accessions | Q14596, Q13173, Q15026, Q96GB6, Q9NRF7 | Gene names | NBR1, 1A13B, KIAA0049, M17S2 | |||
|
Domain Architecture |

|
|||||
| Description | Next to BRCA1 gene 1 protein (Neighbor of BRCA1 gene 1 protein) (Membrane component, chromosome 17, surface marker 2) (1A1-3B). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NBR1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q14596, Q13173, Q15026, Q96GB6, Q9NRF7 | Gene names | NBR1, 1A13B, KIAA0049, M17S2 | |||
|
Domain Architecture |

|
|||||
| Description | Next to BRCA1 gene 1 protein (Neighbor of BRCA1 gene 1 protein) (Membrane component, chromosome 17, surface marker 2) (1A1-3B). | |||||
|
NBR1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.975755 (rank : 2) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P97432, Q9JIH9 | Gene names | Nbr1, M17s2 | |||
|
Domain Architecture |

|
|||||
| Description | Next to BRCA1 gene 1 protein (Neighbor of BRCA1 gene 1 protein) (Membrane component, chromosome 17, surface marker 2). | |||||
|
SQSTM_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 3) | NC score | 0.349450 (rank : 3) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13501, Q13446, Q9BUV7, Q9BVS6, Q9UEU1 | Gene names | SQSTM1, ORCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Phosphotyrosine-independent ligand for the Lck SH2 domain of 62 kDa) (Ubiquitin-binding protein p62) (EBI3-associated protein of 60 kDa) (p60) (EBIAP). | |||||
|
CF106_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 4) | NC score | 0.347409 (rank : 4) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H6K1, Q5VV77, Q96MG5, Q9BUR9 | Gene names | C6orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf106. | |||||
|
SQSTM_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 5) | NC score | 0.340940 (rank : 5) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q64337, Q99JM8 | Gene names | Sqstm1, A170, STAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Ubiquitin-binding protein p62) (STONE14). | |||||
|
CF106_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 6) | NC score | 0.321959 (rank : 6) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3TT38 | Gene names | D17Wsu92e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf106 homolog. | |||||
|
ARHGC_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 7) | NC score | 0.054050 (rank : 22) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
MIB1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 8) | NC score | 0.072758 (rank : 18) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86YT6, Q68D01, Q6YI51, Q8NBY0, Q8TCB5, Q8TCL7, Q9P2M3 | Gene names | MIB1, DIP1, KIAA1323, ZZANK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1) (Zinc finger ZZ type with ankyrin repeat domain protein 2). | |||||
|
MIB1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 9) | NC score | 0.072853 (rank : 17) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80SY4, Q5XK51, Q6IS57, Q6YI52, Q6ZPT8, Q8BNR1, Q8C6W2, Q921Q1 | Gene names | Mib1, Dip1, Kiaa1323, Mib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1). | |||||
|
FGD4_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.031503 (rank : 43) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
HERC2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 11) | NC score | 0.080307 (rank : 14) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.080632 (rank : 13) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
LZTS2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.042493 (rank : 27) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
DTNA_MOUSE
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.109476 (rank : 7) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D2N4, P97319, Q61498, Q61499, Q9QZZ5, Q9WUL9, Q9WUM0 | Gene names | Dtna, Dtn | |||
|
Domain Architecture |
|
|||||
| Description | Dystrobrevin alpha (Alpha-dystrobrevin). | |||||
|
ZSWM2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 15) | NC score | 0.106116 (rank : 8) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D9X6 | Gene names | Zswim2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
AN30A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.029692 (rank : 47) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.037340 (rank : 31) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
MIB2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.072740 (rank : 19) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
MIB2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.073594 (rank : 16) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R516, Q52QU8, Q6PEF6, Q8C1N7, Q8VIB4 | Gene names | Mib2, Skd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Mind bomb-2) (Skeletrophin) (Dystrophin-like protein) (Dyslike). | |||||
|
SNAT_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.061058 (rank : 21) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16613, Q562F4 | Gene names | AANAT, SNAT | |||
|
Domain Architecture |
|
|||||
| Description | Serotonin N-acetyltransferase (EC 2.3.1.87) (Aralkylamine N- acetyltransferase) (AA-NAT) (Serotonin acetylase). | |||||
|
ZO1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.031084 (rank : 45) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ZSWM2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.105663 (rank : 9) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NEG5 | Gene names | ZSWIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
DTNA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.105265 (rank : 10) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y4J8, O15332, O15333, O75697, Q13197, Q13198, Q13199, Q13498, Q13499, Q13500 | Gene names | DTNA, DRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Dystrobrevin-alpha) (Dystrophin-related protein 3). | |||||
|
FANCM_HUMAN
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.034555 (rank : 35) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
FYB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.072508 (rank : 20) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
EZRI_MOUSE
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.022856 (rank : 59) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
FYB_MOUSE
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.076948 (rank : 15) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.033107 (rank : 40) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
DTNB_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.095730 (rank : 11) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60941, O43782, O60881, O75538, Q9UE14, Q9UE15, Q9UE16 | Gene names | DTNB | |||
|
Domain Architecture |
|
|||||
| Description | Dystrobrevin beta (Beta-dystrobrevin) (DTN-B). | |||||
|
SMAP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.027948 (rank : 49) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.024691 (rank : 56) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.017520 (rank : 74) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
DTNB_MOUSE
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.094341 (rank : 12) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70585, O70563, Q9CTZ1 | Gene names | Dtnb | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin beta (Beta-dystrobrevin) (DTN-B) (MDTN-B). | |||||
|
EZRI_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.021101 (rank : 63) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
HIPK4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.005417 (rank : 98) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 799 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NE63 | Gene names | HIPK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeodomain-interacting protein kinase 4 (EC 2.7.11.1). | |||||
|
CCD41_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.020950 (rank : 64) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
DBNL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.034434 (rank : 37) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62418, Q80WP1, Q8BH56 | Gene names | Dbnl, Abp1, Sh3p7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Drebrin-like protein (SH3 domain-containing protein 7) (Actin-binding protein 1). | |||||
|
GRAP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.019621 (rank : 68) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.043136 (rank : 25) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MELPH_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.026598 (rank : 53) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
SF3B2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.037894 (rank : 28) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13435 | Gene names | SF3B2, SAP145 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 2 (Spliceosome-associated protein 145) (SAP 145) (SF3b150) (Pre-mRNA-splicing factor SF3b 145 kDa subunit). | |||||
|
SURF6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.053224 (rank : 23) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75683, Q9BRK9, Q9BTZ5, Q9UK24 | Gene names | SURF6, SURF-6 | |||
|
Domain Architecture |

|
|||||
| Description | Surfeit locus protein 6. | |||||
|
EP300_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.037328 (rank : 32) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
HXC5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.009375 (rank : 93) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00444 | Gene names | HOXC5, HOX3D | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C5 (Hox-3D) (CP11). | |||||
|
HXC5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.009423 (rank : 92) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32043, Q8BJW4 | Gene names | Hoxc5, Hox-3.4, Hoxc-5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C5 (Hox-3.4) (Hox-6.2). | |||||
|
MCA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.024998 (rank : 55) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P31230, Q60659 | Gene names | Scye1, Emap2 | |||
|
Domain Architecture |

|
|||||
| Description | Multisynthetase complex auxiliary component p43 [Contains: Endothelial monocyte-activating polypeptide 2 (EMAP-II) (Small inducible cytokine subfamily E member 1)]. | |||||
|
REN3A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.042619 (rank : 26) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H1J1, Q5T8C3, Q5T8C9, Q7Z6N3, Q86YK1, Q9BZI8 | Gene names | UPF3A, RENT3A, UPF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3A (Nonsense mRNA reducing factor 3A) (Up-frameshift suppressor 3 homolog A) (hUpf3). | |||||
|
RILP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.026666 (rank : 52) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96NA2, Q71RE6, Q9BSL3, Q9BYS3 | Gene names | RILP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab-interacting lysosomal protein. | |||||
|
SMBP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.031449 (rank : 44) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
TCEA2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.022178 (rank : 61) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QVN7, O08667 | Gene names | Tcea2 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription elongation factor A protein 2 (Transcription elongation factor S-II protein 2) (Testis-specific S-II) (Protein S-II-T1) (Transcription elongation factor TFIIS.l). | |||||
|
ZBP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.037601 (rank : 30) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QY24, Q8VE02, Q9D8B9 | Gene names | Zbp1, Dlm1 | |||
|
Domain Architecture |
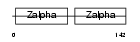
|
|||||
| Description | Z-DNA-binding protein 1 (Tumor stroma and activated macrophage protein DLM-1). | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.032263 (rank : 41) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
DRG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.034576 (rank : 33) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y295, Q6FGP8, Q8WW69, Q9UGF2 | Gene names | DRG1 | |||
|
Domain Architecture |
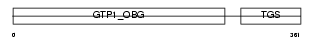
|
|||||
| Description | Developmentally-regulated GTP-binding protein 1 (DRG 1). | |||||
|
DRG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.034562 (rank : 34) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P32233 | Gene names | Drg1, Drg, Nedd-3, Nedd3 | |||
|
Domain Architecture |
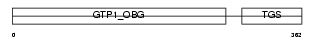
|
|||||
| Description | Developmentally-regulated GTP-binding protein 1 (DRG 1) (Protein NEDD3) (Neural precursor cell expressed developmentally down-regulated protein 3). | |||||
|
FNDC3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.014469 (rank : 84) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BX90, Q2VEY7, Q6ZQ15, Q811D3, Q8BTM3 | Gene names | Fndc3a, D14Ertd453e, Fndc3, Kiaa0970 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type-III domain-containing protein 3a. | |||||
|
KPCD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.002577 (rank : 101) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15139 | Gene names | PRKD1, PKD, PKD1, PRKCM | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase D1 (EC 2.7.11.13) (nPKC-D1) (Protein kinase D) (Protein kinase C mu type) (nPKC-mu). | |||||
|
KPCZ_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.002077 (rank : 105) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02956 | Gene names | Prkcz, Pkcz | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C zeta type (EC 2.7.11.13) (nPKC-zeta). | |||||
|
LZTS2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.029953 (rank : 46) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BRK4, Q8N3I0, Q96J79, Q96JL2 | Gene names | LZTS2, KIAA1813, LAPSER1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2 (Protein LAPSER1). | |||||
|
M3K13_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.002308 (rank : 102) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43283 | Gene names | MAP3K13, LZK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 13 (EC 2.7.11.25) (Mixed lineage kinase) (MLK) (Leucine zipper-bearing kinase). | |||||
|
MMRN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.015656 (rank : 80) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13201, Q6P3T8 | Gene names | MMRN1, ECM, EMILIN4, MMRN | |||
|
Domain Architecture |

|
|||||
| Description | Multimerin-1 precursor (Endothelial cell multimerin 1) (EMILIN-4) (Elastin microfibril interface located protein 4) (Elastin microfibril interfacer 4). | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.002223 (rank : 104) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
ZO1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.024658 (rank : 57) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.031916 (rank : 42) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.034465 (rank : 36) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.034090 (rank : 38) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.019387 (rank : 69) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.021162 (rank : 62) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
PRGR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.006231 (rank : 97) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06401, Q9UPF7 | Gene names | PGR, NR3C3 | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
RPGR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.027776 (rank : 50) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
SELS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.045309 (rank : 24) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BCZ4, Q921S1, Q9DB55 | Gene names | Sels, H47, Vimp | |||
|
Domain Architecture |

|
|||||
| Description | Selenoprotein S (VCP-interacting membrane protein) (Minor histocompatibility antigen H47). | |||||
|
TEX14_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.014118 (rank : 85) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.033150 (rank : 39) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.018194 (rank : 73) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CAH8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.006947 (rank : 95) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35219 | Gene names | CA8, CALS | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase-related protein (CARP) (CA-VIII). | |||||
|
CCD25_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.020668 (rank : 65) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q78PG9, Q9CSQ8, Q9CUF8 | Gene names | Ccdc25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 25. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.027645 (rank : 51) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
NAF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.015157 (rank : 81) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.002263 (rank : 103) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
NRBF2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.025872 (rank : 54) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96F24, Q86UR2, Q96NP6, Q9H0S9, Q9H2I2 | Gene names | NRBF2, COPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding factor 2 (NRBF-2) (Comodulator of PPAR and RXR). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.011815 (rank : 89) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RNF17_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.016053 (rank : 79) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MV7 | Gene names | Rnf17 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 17. | |||||
|
UBXD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.016846 (rank : 76) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92575, Q8IYM5 | Gene names | UBXD2, KIAA0242, UBXDC1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
ABCBB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.003810 (rank : 99) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QY30, Q9QZE8 | Gene names | Abcb11, Bsep, Spgp | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt export pump (ATP-binding cassette sub-family B member 11) (Sister of P-glycoprotein). | |||||
|
ASPP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.018998 (rank : 71) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.018898 (rank : 72) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
DC1I1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.020167 (rank : 66) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14576, Q9Y2X1 | Gene names | DYNC1I1, DNCI1, DNCIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic dynein 1 intermediate chain 1 (Dynein intermediate chain 1, cytosolic) (DH IC-1) (Cytoplasmic dynein intermediate chain 1). | |||||
|
DC1I1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.029333 (rank : 48) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88485, O88486 | Gene names | Dync1i1, Dnci1, Dncic1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic dynein 1 intermediate chain 1 (Dynein intermediate chain 1, cytosolic) (DH IC-1) (Cytoplasmic dynein intermediate chain 1). | |||||
|
DC1I2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.019914 (rank : 67) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13409, Q96NG7, Q96S87, Q9BXZ5, Q9NT58 | Gene names | DYNC1I2, DNCI2, DNCIC2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic dynein 1 intermediate chain 2 (Dynein intermediate chain 2, cytosolic) (DH IC-2) (Cytoplasmic dynein intermediate chain 2). | |||||
|
DC1I2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.019191 (rank : 70) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88487, Q3TGH7 | Gene names | Dync1i2, Dnci2, Dncic2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic dynein 1 intermediate chain 2 (Dynein intermediate chain 2, cytosolic) (DH IC-2) (Cytoplasmic dynein intermediate chain 2). | |||||
|
DNJC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.008639 (rank : 94) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96KC8 | Gene names | DNAJC1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1. | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.014063 (rank : 86) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.016193 (rank : 78) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.017208 (rank : 75) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MORC4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.013481 (rank : 87) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TE76, Q5JUK7, Q96MZ2, Q9HAI7 | Gene names | MORC4, ZCWCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
MORC4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.014672 (rank : 82) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BMD7, Q4KMM6, Q8BX95, Q9CS96 | Gene names | Morc4, Zcwcc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
MTG8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.010185 (rank : 90) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61909 | Gene names | Runx1t1, Cbfa2t1, Cbfa2t1h, Mtg8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.003656 (rank : 100) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
MYOM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.006261 (rank : 96) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62234 | Gene names | Myom1 | |||
|
Domain Architecture |

|
|||||
| Description | Myomesin-1 (Skelemin). | |||||
|
NRBF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.023108 (rank : 58) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCQ3, Q9DCG3 | Gene names | Nrbf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding factor 2 (NRBF-2). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.022739 (rank : 60) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
SC24D_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.012167 (rank : 88) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94855 | Gene names | SEC24D, KIAA0755 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24D (SEC24-related protein D). | |||||
|
SSF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.014565 (rank : 83) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YU8 | Gene names | Ppan | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of SWI4 1 homolog (Ssf-1) (Peter Pan homolog). | |||||
|
TBC12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.009429 (rank : 91) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60347, Q5VYA6, Q8WX26, Q8WX59, Q9UG83 | Gene names | TBC1D12, KIAA0608 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 12. | |||||
|
TCEA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.016651 (rank : 77) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15560, Q8TD37, Q8TD38 | Gene names | TCEA2 | |||
|
Domain Architecture |
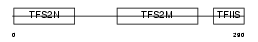
|
|||||
| Description | Transcription elongation factor A protein 2 (Transcription elongation factor S-II protein 2) (Testis-specific S-II) (Transcription elongation factor TFIIS.l). | |||||
|
ZBP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.037665 (rank : 29) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H171, Q5JY39, Q9BYW4 | Gene names | ZBP1, C20orf183, DLM1 | |||
|
Domain Architecture |
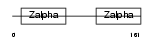
|
|||||
| Description | Z-DNA-binding protein 1 (Tumor stroma and activated macrophage protein DLM-1). | |||||
|
NBR1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q14596, Q13173, Q15026, Q96GB6, Q9NRF7 | Gene names | NBR1, 1A13B, KIAA0049, M17S2 | |||
|
Domain Architecture |

|
|||||
| Description | Next to BRCA1 gene 1 protein (Neighbor of BRCA1 gene 1 protein) (Membrane component, chromosome 17, surface marker 2) (1A1-3B). | |||||
|
NBR1_MOUSE
|
||||||
| NC score | 0.975755 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P97432, Q9JIH9 | Gene names | Nbr1, M17s2 | |||
|
Domain Architecture |

|
|||||
| Description | Next to BRCA1 gene 1 protein (Neighbor of BRCA1 gene 1 protein) (Membrane component, chromosome 17, surface marker 2). | |||||
|
SQSTM_HUMAN
|
||||||
| NC score | 0.349450 (rank : 3) | θ value | 1.53129e-07 (rank : 3) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13501, Q13446, Q9BUV7, Q9BVS6, Q9UEU1 | Gene names | SQSTM1, ORCA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Phosphotyrosine-independent ligand for the Lck SH2 domain of 62 kDa) (Ubiquitin-binding protein p62) (EBI3-associated protein of 60 kDa) (p60) (EBIAP). | |||||
|
CF106_HUMAN
|
||||||
| NC score | 0.347409 (rank : 4) | θ value | 2.61198e-07 (rank : 4) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H6K1, Q5VV77, Q96MG5, Q9BUR9 | Gene names | C6orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf106. | |||||
|
SQSTM_MOUSE
|
||||||
| NC score | 0.340940 (rank : 5) | θ value | 2.61198e-07 (rank : 5) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q64337, Q99JM8 | Gene names | Sqstm1, A170, STAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sequestosome-1 (Ubiquitin-binding protein p62) (STONE14). | |||||
|
CF106_MOUSE
|
||||||
| NC score | 0.321959 (rank : 6) | θ value | 1.43324e-05 (rank : 6) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3TT38 | Gene names | D17Wsu92e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf106 homolog. | |||||
|
DTNA_MOUSE
|
||||||
| NC score | 0.109476 (rank : 7) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D2N4, P97319, Q61498, Q61499, Q9QZZ5, Q9WUL9, Q9WUM0 | Gene names | Dtna, Dtn | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Alpha-dystrobrevin). | |||||
|
ZSWM2_MOUSE
|
||||||
| NC score | 0.106116 (rank : 8) | θ value | 0.279714 (rank : 15) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D9X6 | Gene names | Zswim2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
ZSWM2_HUMAN
|
||||||
| NC score | 0.105663 (rank : 9) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NEG5 | Gene names | ZSWIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
DTNA_HUMAN
|
||||||
| NC score | 0.105265 (rank : 10) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y4J8, O15332, O15333, O75697, Q13197, Q13198, Q13199, Q13498, Q13499, Q13500 | Gene names | DTNA, DRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Dystrobrevin-alpha) (Dystrophin-related protein 3). | |||||
|
DTNB_HUMAN
|
||||||
| NC score | 0.095730 (rank : 11) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60941, O43782, O60881, O75538, Q9UE14, Q9UE15, Q9UE16 | Gene names | DTNB | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin beta (Beta-dystrobrevin) (DTN-B). | |||||
|
DTNB_MOUSE
|
||||||
| NC score | 0.094341 (rank : 12) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70585, O70563, Q9CTZ1 | Gene names | Dtnb | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin beta (Beta-dystrobrevin) (DTN-B) (MDTN-B). | |||||
|
HERC2_MOUSE
|
||||||
| NC score | 0.080632 (rank : 13) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_HUMAN
|
||||||
| NC score | 0.080307 (rank : 14) | θ value | 0.0961366 (rank : 11) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
FYB_MOUSE
|
||||||
| NC score | 0.076948 (rank : 15) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
MIB2_MOUSE
|
||||||
| NC score | 0.073594 (rank : 16) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R516, Q52QU8, Q6PEF6, Q8C1N7, Q8VIB4 | Gene names | Mib2, Skd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Mind bomb-2) (Skeletrophin) (Dystrophin-like protein) (Dyslike). | |||||
|
MIB1_MOUSE
|
||||||
| NC score | 0.072853 (rank : 17) | θ value | 0.0330416 (rank : 9) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80SY4, Q5XK51, Q6IS57, Q6YI52, Q6ZPT8, Q8BNR1, Q8C6W2, Q921Q1 | Gene names | Mib1, Dip1, Kiaa1323, Mib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1). | |||||
|
MIB1_HUMAN
|
||||||
| NC score | 0.072758 (rank : 18) | θ value | 0.0330416 (rank : 8) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86YT6, Q68D01, Q6YI51, Q8NBY0, Q8TCB5, Q8TCL7, Q9P2M3 | Gene names | MIB1, DIP1, KIAA1323, ZZANK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1) (Zinc finger ZZ type with ankyrin repeat domain protein 2). | |||||
|
MIB2_HUMAN
|
||||||
| NC score | 0.072740 (rank : 19) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
FYB_HUMAN
|
||||||
| NC score | 0.072508 (rank : 20) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
SNAT_HUMAN
|
||||||
| NC score | 0.061058 (rank : 21) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16613, Q562F4 | Gene names | AANAT, SNAT | |||
|
Domain Architecture |

|
|||||
| Description | Serotonin N-acetyltransferase (EC 2.3.1.87) (Aralkylamine N- acetyltransferase) (AA-NAT) (Serotonin acetylase). | |||||
|
ARHGC_HUMAN
|
||||||
| NC score | 0.054050 (rank : 22) | θ value | 0.0330416 (rank : 7) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
SURF6_HUMAN
|
||||||
| NC score | 0.053224 (rank : 23) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75683, Q9BRK9, Q9BTZ5, Q9UK24 | Gene names | SURF6, SURF-6 | |||
|
Domain Architecture |

|
|||||
| Description | Surfeit locus protein 6. | |||||
|
SELS_MOUSE
|
||||||
| NC score | 0.045309 (rank : 24) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BCZ4, Q921S1, Q9DB55 | Gene names | Sels, H47, Vimp | |||
|
Domain Architecture |

|
|||||
| Description | Selenoprotein S (VCP-interacting membrane protein) (Minor histocompatibility antigen H47). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.043136 (rank : 25) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
REN3A_HUMAN
|
||||||
| NC score | 0.042619 (rank : 26) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H1J1, Q5T8C3, Q5T8C9, Q7Z6N3, Q86YK1, Q9BZI8 | Gene names | UPF3A, RENT3A, UPF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3A (Nonsense mRNA reducing factor 3A) (Up-frameshift suppressor 3 homolog A) (hUpf3). | |||||
|
LZTS2_MOUSE
|
||||||
| NC score | 0.042493 (rank : 27) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
SF3B2_HUMAN
|
||||||
| NC score | 0.037894 (rank : 28) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13435 | Gene names | SF3B2, SAP145 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 2 (Spliceosome-associated protein 145) (SAP 145) (SF3b150) (Pre-mRNA-splicing factor SF3b 145 kDa subunit). | |||||
|
ZBP1_HUMAN
|
||||||
| NC score | 0.037665 (rank : 29) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H171, Q5JY39, Q9BYW4 | Gene names | ZBP1, C20orf183, DLM1 | |||
|
Domain Architecture |

|
|||||
| Description | Z-DNA-binding protein 1 (Tumor stroma and activated macrophage protein DLM-1). | |||||
|
ZBP1_MOUSE
|
||||||
| NC score | 0.037601 (rank : 30) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QY24, Q8VE02, Q9D8B9 | Gene names | Zbp1, Dlm1 | |||
|
Domain Architecture |

|
|||||
| Description | Z-DNA-binding protein 1 (Tumor stroma and activated macrophage protein DLM-1). | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.037340 (rank : 31) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.037328 (rank : 32) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
DRG1_HUMAN
|
||||||
| NC score | 0.034576 (rank : 33) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y295, Q6FGP8, Q8WW69, Q9UGF2 | Gene names | DRG1 | |||
|
Domain Architecture |

|
|||||
| Description | Developmentally-regulated GTP-binding protein 1 (DRG 1). | |||||
|
DRG1_MOUSE
|
||||||
| NC score | 0.034562 (rank : 34) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P32233 | Gene names | Drg1, Drg, Nedd-3, Nedd3 | |||
|
Domain Architecture |

|
|||||
| Description | Developmentally-regulated GTP-binding protein 1 (DRG 1) (Protein NEDD3) (Neural precursor cell expressed developmentally down-regulated protein 3). | |||||
|
FANCM_HUMAN
|
||||||
| NC score | 0.034555 (rank : 35) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.034465 (rank : 36) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
DBNL_MOUSE
|
||||||
| NC score | 0.034434 (rank : 37) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62418, Q80WP1, Q8BH56 | Gene names | Dbnl, Abp1, Sh3p7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Drebrin-like protein (SH3 domain-containing protein 7) (Actin-binding protein 1). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.034090 (rank : 38) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.033150 (rank : 39) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.033107 (rank : 40) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.032263 (rank : 41) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.031916 (rank : 42) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
FGD4_MOUSE
|
||||||
| NC score | 0.031503 (rank : 43) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
SMBP2_HUMAN
|
||||||
| NC score | 0.031449 (rank : 44) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.031084 (rank : 45) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
LZTS2_HUMAN
|
||||||
| NC score | 0.029953 (rank : 46) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BRK4, Q8N3I0, Q96J79, Q96JL2 | Gene names | LZTS2, KIAA1813, LAPSER1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2 (Protein LAPSER1). | |||||
|
AN30A_HUMAN
|
||||||
| NC score | 0.029692 (rank : 47) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1119 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BXX3, Q5W025 | Gene names | ANKRD30A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 30A (NY-BR-1 antigen). | |||||
|
DC1I1_MOUSE
|
||||||
| NC score | 0.029333 (rank : 48) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88485, O88486 | Gene names | Dync1i1, Dnci1, Dncic1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic dynein 1 intermediate chain 1 (Dynein intermediate chain 1, cytosolic) (DH IC-1) (Cytoplasmic dynein intermediate chain 1). | |||||
|
SMAP1_HUMAN
|
||||||
| NC score | 0.027948 (rank : 49) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IYB5, Q53H70, Q5SYQ2, Q6PK24, Q8NDH4, Q96L38, Q96L39, Q9H8X4 | Gene names | SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
RPGR_MOUSE
|
||||||
| NC score | 0.027776 (rank : 50) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.027645 (rank : 51) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
RILP_HUMAN
|
||||||
| NC score | 0.026666 (rank : 52) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96NA2, Q71RE6, Q9BSL3, Q9BYS3 | Gene names | RILP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab-interacting lysosomal protein. | |||||
|
MELPH_MOUSE
|
||||||
| NC score | 0.026598 (rank : 53) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
NRBF2_HUMAN
|
||||||
| NC score | 0.025872 (rank : 54) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96F24, Q86UR2, Q96NP6, Q9H0S9, Q9H2I2 | Gene names | NRBF2, COPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding factor 2 (NRBF-2) (Comodulator of PPAR and RXR). | |||||
|
MCA1_MOUSE
|
||||||
| NC score | 0.024998 (rank : 55) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P31230, Q60659 | Gene names | Scye1, Emap2 | |||
|
Domain Architecture |

|
|||||
| Description | Multisynthetase complex auxiliary component p43 [Contains: Endothelial monocyte-activating polypeptide 2 (EMAP-II) (Small inducible cytokine subfamily E member 1)]. | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.024691 (rank : 56) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
ZO1_MOUSE
|
||||||
| NC score | 0.024658 (rank : 57) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
NRBF2_MOUSE
|
||||||
| NC score | 0.023108 (rank : 58) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCQ3, Q9DCG3 | Gene names | Nrbf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-binding factor 2 (NRBF-2). | |||||
|
EZRI_MOUSE
|
||||||
| NC score | 0.022856 (rank : 59) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26040, Q80ZT8, Q9DCI1 | Gene names | Vil2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.022739 (rank : 60) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
TCEA2_MOUSE
|
||||||
| NC score | 0.022178 (rank : 61) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QVN7, O08667 | Gene names | Tcea2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor A protein 2 (Transcription elongation factor S-II protein 2) (Testis-specific S-II) (Protein S-II-T1) (Transcription elongation factor TFIIS.l). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.021162 (rank : 62) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
EZRI_HUMAN
|
||||||
| NC score | 0.021101 (rank : 63) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 663 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15311, P23714, Q96CU8, Q9NSJ4 | Gene names | VIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin (p81) (Cytovillin) (Villin-2). | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.020950 (rank : 64) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
CCD25_MOUSE
|
||||||
| NC score | 0.020668 (rank : 65) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q78PG9, Q9CSQ8, Q9CUF8 | Gene names | Ccdc25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 25. | |||||
|
DC1I1_HUMAN
|
||||||
| NC score | 0.020167 (rank : 66) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14576, Q9Y2X1 | Gene names | DYNC1I1, DNCI1, DNCIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic dynein 1 intermediate chain 1 (Dynein intermediate chain 1, cytosolic) (DH IC-1) (Cytoplasmic dynein intermediate chain 1). | |||||
|
DC1I2_HUMAN
|
||||||
| NC score | 0.019914 (rank : 67) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13409, Q96NG7, Q96S87, Q9BXZ5, Q9NT58 | Gene names | DYNC1I2, DNCI2, DNCIC2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic dynein 1 intermediate chain 2 (Dynein intermediate chain 2, cytosolic) (DH IC-2) (Cytoplasmic dynein intermediate chain 2). | |||||
|
GRAP1_HUMAN
|
||||||
| NC score | 0.019621 (rank : 68) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.019387 (rank : 69) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
DC1I2_MOUSE
|
||||||
| NC score | 0.019191 (rank : 70) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88487, Q3TGH7 | Gene names | Dync1i2, Dnci2, Dncic2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic dynein 1 intermediate chain 2 (Dynein intermediate chain 2, cytosolic) (DH IC-2) (Cytoplasmic dynein intermediate chain 2). | |||||
|
ASPP1_HUMAN
|
||||||
| NC score | 0.018998 (rank : 71) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.018898 (rank : 72) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.018194 (rank : 73) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.017520 (rank : 74) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.017208 (rank : 75) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
UBXD2_HUMAN
|
||||||
| NC score | 0.016846 (rank : 76) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92575, Q8IYM5 | Gene names | UBXD2, KIAA0242, UBXDC1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
TCEA2_HUMAN
|
||||||
| NC score | 0.016651 (rank : 77) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15560, Q8TD37, Q8TD38 | Gene names | TCEA2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor A protein 2 (Transcription elongation factor S-II protein 2) (Testis-specific S-II) (Transcription elongation factor TFIIS.l). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.016193 (rank : 78) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
RNF17_MOUSE
|
||||||
| NC score | 0.016053 (rank : 79) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MV7 | Gene names | Rnf17 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 17. | |||||
|
MMRN1_HUMAN
|
||||||
| NC score | 0.015656 (rank : 80) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13201, Q6P3T8 | Gene names | MMRN1, ECM, EMILIN4, MMRN | |||
|
Domain Architecture |

|
|||||
| Description | Multimerin-1 precursor (Endothelial cell multimerin 1) (EMILIN-4) (Elastin microfibril interface located protein 4) (Elastin microfibril interfacer 4). | |||||
|
NAF1_MOUSE
|
||||||
| NC score | 0.015157 (rank : 81) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
MORC4_MOUSE
|
||||||
| NC score | 0.014672 (rank : 82) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BMD7, Q4KMM6, Q8BX95, Q9CS96 | Gene names | Morc4, Zcwcc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
SSF1_MOUSE
|
||||||
| NC score | 0.014565 (rank : 83) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YU8 | Gene names | Ppan | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of SWI4 1 homolog (Ssf-1) (Peter Pan homolog). | |||||
|
FNDC3_MOUSE
|
||||||
| NC score | 0.014469 (rank : 84) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BX90, Q2VEY7, Q6ZQ15, Q811D3, Q8BTM3 | Gene names | Fndc3a, D14Ertd453e, Fndc3, Kiaa0970 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type-III domain-containing protein 3a. | |||||
|
TEX14_MOUSE
|
||||||
| NC score | 0.014118 (rank : 85) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | 0.014063 (rank : 86) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
MORC4_HUMAN
|
||||||
| NC score | 0.013481 (rank : 87) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TE76, Q5JUK7, Q96MZ2, Q9HAI7 | Gene names | MORC4, ZCWCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
SC24D_HUMAN
|
||||||
| NC score | 0.012167 (rank : 88) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94855 | Gene names | SEC24D, KIAA0755 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24D (SEC24-related protein D). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.011815 (rank : 89) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MTG8_MOUSE
|
||||||
| NC score | 0.010185 (rank : 90) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61909 | Gene names | Runx1t1, Cbfa2t1, Cbfa2t1h, Mtg8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T1 (Protein MTG8). | |||||
|
TBC12_HUMAN
|
||||||
| NC score | 0.009429 (rank : 91) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60347, Q5VYA6, Q8WX26, Q8WX59, Q9UG83 | Gene names | TBC1D12, KIAA0608 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 12. | |||||
|
HXC5_MOUSE
|
||||||
| NC score | 0.009423 (rank : 92) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32043, Q8BJW4 | Gene names | Hoxc5, Hox-3.4, Hoxc-5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C5 (Hox-3.4) (Hox-6.2). | |||||
|
HXC5_HUMAN
|
||||||
| NC score | 0.009375 (rank : 93) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00444 | Gene names | HOXC5, HOX3D | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-C5 (Hox-3D) (CP11). | |||||
|
DNJC1_HUMAN
|
||||||
| NC score | 0.008639 (rank : 94) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96KC8 | Gene names | DNAJC1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 1. | |||||
|
CAH8_HUMAN
|
||||||
| NC score | 0.006947 (rank : 95) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35219 | Gene names | CA8, CALS | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase-related protein (CARP) (CA-VIII). | |||||
|
MYOM1_MOUSE
|
||||||
| NC score | 0.006261 (rank : 96) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62234 | Gene names | Myom1 | |||
|
Domain Architecture |

|
|||||
| Description | Myomesin-1 (Skelemin). | |||||
|
PRGR_HUMAN
|
||||||
| NC score | 0.006231 (rank : 97) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06401, Q9UPF7 | Gene names | PGR, NR3C3 | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
HIPK4_HUMAN
|
||||||
| NC score | 0.005417 (rank : 98) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 799 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NE63 | Gene names | HIPK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeodomain-interacting protein kinase 4 (EC 2.7.11.1). | |||||
|
ABCBB_MOUSE
|
||||||
| NC score | 0.003810 (rank : 99) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QY30, Q9QZE8 | Gene names | Abcb11, Bsep, Spgp | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt export pump (ATP-binding cassette sub-family B member 11) (Sister of P-glycoprotein). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.003656 (rank : 100) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
KPCD1_HUMAN
|
||||||
| NC score | 0.002577 (rank : 101) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15139 | Gene names | PRKD1, PKD, PKD1, PRKCM | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase D1 (EC 2.7.11.13) (nPKC-D1) (Protein kinase D) (Protein kinase C mu type) (nPKC-mu). | |||||
|
M3K13_HUMAN
|
||||||
| NC score | 0.002308 (rank : 102) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43283 | Gene names | MAP3K13, LZK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 13 (EC 2.7.11.25) (Mixed lineage kinase) (MLK) (Leucine zipper-bearing kinase). | |||||
|
NEK1_MOUSE
|
||||||
| NC score | 0.002263 (rank : 103) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.002223 (rank : 104) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
KPCZ_MOUSE
|
||||||
| NC score | 0.002077 (rank : 105) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 105 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02956 | Gene names | Prkcz, Pkcz | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C zeta type (EC 2.7.11.13) (nPKC-zeta). | |||||