Please be patient as the page loads
|
K1143_MOUSE
|
||||||
| SwissProt Accessions | Q8K039, Q9CSR4, Q9D0W8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1143 homolog. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
K1143_MOUSE
|
||||||
| θ value | 1.31072e-83 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K039, Q9CSR4, Q9D0W8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1143 homolog. | |||||
|
K1143_HUMAN
|
||||||
| θ value | 6.7473e-72 (rank : 2) | NC score | 0.970230 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96AT1, Q96HJ8, Q9ULS7 | Gene names | KIAA1143 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1143. | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 3) | NC score | 0.098052 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 4) | NC score | 0.099299 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 5) | NC score | 0.047708 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
CHD6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 6) | NC score | 0.030610 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
DKC1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 7) | NC score | 0.069334 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60832, O43845, Q96G67, Q9Y505 | Gene names | DKC1, NOLA4 | |||
|
Domain Architecture |
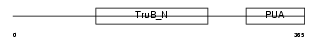
|
|||||
| Description | H/ACA ribonucleoprotein complex subunit 4 (EC 5.4.99.-) (Dyskerin) (Nucleolar protein family A member 4) (snoRNP protein DKC1) (Nopp140- associated protein of 57 kDa) (Nucleolar protein NAP57) (CBF5 homolog). | |||||
|
ZCPW1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 8) | NC score | 0.103443 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 9) | NC score | 0.037370 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 10) | NC score | 0.074218 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TPM2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.047985 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P58774, P02560, P46901 | Gene names | Tpm2, Tpm-2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.062055 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
TPM2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 13) | NC score | 0.046338 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 677 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P07951, P06468, Q13894, Q53FM4, Q5TCU4, Q5TCU7, Q9UH67 | Gene names | TPM2, TMSB | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
E41L2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.039954 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
CALX_HUMAN
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.035499 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
CJ118_MOUSE
|
||||||
| θ value | 1.81305 (rank : 16) | NC score | 0.024387 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.072692 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
HSP74_MOUSE
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.024041 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.067535 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
TPM3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.042375 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P06753, P12324, Q969Q2, Q9NQH8 | Gene names | TPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma) (hTM5). | |||||
|
DEK_MOUSE
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.055222 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
E41L1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.043548 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.043297 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z2H5 | Gene names | Epb41l1, Epb4, Epb4.1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.021971 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ZFP91_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.009968 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JP5, Q86V47, Q96JP4, Q96QA3 | Gene names | ZFP91 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 homolog (Zfp-91). | |||||
|
ZFP91_MOUSE
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.009788 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62511, Q8BPY3, Q8C2B4 | Gene names | Zfp91, Pzf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 (Zfp-91) (Zinc finger protein PZF) (Penta Zf protein). | |||||
|
CIR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.048762 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
INADL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.016008 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
TULP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.029529 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z273 | Gene names | Tulp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
CKAP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.047603 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3V1H1, Q66LN2, Q8BSF0 | Gene names | Ckap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 2. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.068529 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LYAR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.035532 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08288, Q9D9X2 | Gene names | Lyar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.038507 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
BICD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.026914 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
IF38_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.056225 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99613, O00215 | Gene names | EIF3S8 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 8 (eIF3 p110) (eIF3c). | |||||
|
RAD52_MOUSE
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.030922 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43352 | Gene names | Rad52 | |||
|
Domain Architecture |
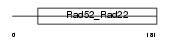
|
|||||
| Description | DNA repair protein RAD52 homolog. | |||||
|
SSRP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.026451 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
TPM3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.039411 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P21107, Q09021, Q60606, Q80SW6, Q9EPW3 | Gene names | Tpm3, Tpm-5, Tpm5 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma). | |||||
|
BICD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.028513 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
CJ006_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.026637 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IX21, Q5W0L8, Q9NPE8 | Gene names | C10orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf6. | |||||
|
HRX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.025447 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
PRP4B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.004343 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.032104 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
ANKS3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.005915 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CZK6, Q6ZPF6, Q80X46 | Gene names | Anks3, Kiaa1977 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 3. | |||||
|
CADH3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.003476 (rank : 51) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22223 | Gene names | CDH3, CDHP | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-3 precursor (Placental-cadherin) (P-cadherin). | |||||
|
DYXC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.015286 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R368 | Gene names | Dyx1c1, Ekn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein homolog. | |||||
|
ITPR2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.011194 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z329, P70226, Q5DWM3, Q5DWM5, Q61744, Q8R3B0 | Gene names | Itpr2, Itpr5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2) (Inositol 1,4,5-trisphosphate type V receptor). | |||||
|
TR150_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.043878 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
NUCKS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.050992 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.051909 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.051230 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
K1143_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.31072e-83 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K039, Q9CSR4, Q9D0W8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1143 homolog. | |||||
|
K1143_HUMAN
|
||||||
| NC score | 0.970230 (rank : 2) | θ value | 6.7473e-72 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96AT1, Q96HJ8, Q9ULS7 | Gene names | KIAA1143 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1143. | |||||
|
ZCPW1_MOUSE
|
||||||
| NC score | 0.103443 (rank : 3) | θ value | 0.365318 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.099299 (rank : 4) | θ value | 0.279714 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.098052 (rank : 5) | θ value | 0.0563607 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.074218 (rank : 6) | θ value | 0.813845 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.072692 (rank : 7) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
DKC1_HUMAN
|
||||||
| NC score | 0.069334 (rank : 8) | θ value | 0.365318 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60832, O43845, Q96G67, Q9Y505 | Gene names | DKC1, NOLA4 | |||
|
Domain Architecture |

|
|||||
| Description | H/ACA ribonucleoprotein complex subunit 4 (EC 5.4.99.-) (Dyskerin) (Nucleolar protein family A member 4) (snoRNP protein DKC1) (Nopp140- associated protein of 57 kDa) (Nucleolar protein NAP57) (CBF5 homolog). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.068529 (rank : 9) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.067535 (rank : 10) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.062055 (rank : 11) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
IF38_HUMAN
|
||||||
| NC score | 0.056225 (rank : 12) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99613, O00215 | Gene names | EIF3S8 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 8 (eIF3 p110) (eIF3c). | |||||
|
DEK_MOUSE
|
||||||
| NC score | 0.055222 (rank : 13) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.051909 (rank : 14) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.051230 (rank : 15) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
NUCKS_MOUSE
|
||||||
| NC score | 0.050992 (rank : 16) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
CIR_HUMAN
|
||||||
| NC score | 0.048762 (rank : 17) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
TPM2_MOUSE
|
||||||
| NC score | 0.047985 (rank : 18) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P58774, P02560, P46901 | Gene names | Tpm2, Tpm-2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.047708 (rank : 19) | θ value | 0.279714 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
CKAP2_MOUSE
|
||||||
| NC score | 0.047603 (rank : 20) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3V1H1, Q66LN2, Q8BSF0 | Gene names | Ckap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 2. | |||||
|
TPM2_HUMAN
|
||||||
| NC score | 0.046338 (rank : 21) | θ value | 1.06291 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 677 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P07951, P06468, Q13894, Q53FM4, Q5TCU4, Q5TCU7, Q9UH67 | Gene names | TPM2, TMSB | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.043878 (rank : 22) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
E41L1_HUMAN
|
||||||
| NC score | 0.043548 (rank : 23) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
E41L1_MOUSE
|
||||||
| NC score | 0.043297 (rank : 24) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z2H5 | Gene names | Epb41l1, Epb4, Epb4.1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
TPM3_HUMAN
|
||||||
| NC score | 0.042375 (rank : 25) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P06753, P12324, Q969Q2, Q9NQH8 | Gene names | TPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma) (hTM5). | |||||
|
E41L2_HUMAN
|
||||||
| NC score | 0.039954 (rank : 26) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
TPM3_MOUSE
|
||||||
| NC score | 0.039411 (rank : 27) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P21107, Q09021, Q60606, Q80SW6, Q9EPW3 | Gene names | Tpm3, Tpm-5, Tpm5 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma). | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.038507 (rank : 28) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.037370 (rank : 29) | θ value | 0.813845 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
LYAR_MOUSE
|
||||||
| NC score | 0.035532 (rank : 30) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08288, Q9D9X2 | Gene names | Lyar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth-regulating nucleolar protein. | |||||
|
CALX_HUMAN
|
||||||
| NC score | 0.035499 (rank : 31) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.032104 (rank : 32) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
RAD52_MOUSE
|
||||||
| NC score | 0.030922 (rank : 33) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43352 | Gene names | Rad52 | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair protein RAD52 homolog. | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.030610 (rank : 34) | θ value | 0.365318 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
TULP1_MOUSE
|
||||||
| NC score | 0.029529 (rank : 35) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z273 | Gene names | Tulp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
BICD1_MOUSE
|
||||||
| NC score | 0.028513 (rank : 36) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
BICD1_HUMAN
|
||||||
| NC score | 0.026914 (rank : 37) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
CJ006_HUMAN
|
||||||
| NC score | 0.026637 (rank : 38) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IX21, Q5W0L8, Q9NPE8 | Gene names | C10orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf6. | |||||
|
SSRP1_MOUSE
|
||||||
| NC score | 0.026451 (rank : 39) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.025447 (rank : 40) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
CJ118_MOUSE
|
||||||
| NC score | 0.024387 (rank : 41) | θ value | 1.81305 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
HSP74_MOUSE
|
||||||
| NC score | 0.024041 (rank : 42) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.021971 (rank : 43) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
INADL_HUMAN
|
||||||
| NC score | 0.016008 (rank : 44) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
DYXC1_MOUSE
|
||||||
| NC score | 0.015286 (rank : 45) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R368 | Gene names | Dyx1c1, Ekn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein homolog. | |||||
|
ITPR2_MOUSE
|
||||||
| NC score | 0.011194 (rank : 46) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z329, P70226, Q5DWM3, Q5DWM5, Q61744, Q8R3B0 | Gene names | Itpr2, Itpr5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2) (Inositol 1,4,5-trisphosphate type V receptor). | |||||
|
ZFP91_HUMAN
|
||||||
| NC score | 0.009968 (rank : 47) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JP5, Q86V47, Q96JP4, Q96QA3 | Gene names | ZFP91 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 homolog (Zfp-91). | |||||
|
ZFP91_MOUSE
|
||||||
| NC score | 0.009788 (rank : 48) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62511, Q8BPY3, Q8C2B4 | Gene names | Zfp91, Pzf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 (Zfp-91) (Zinc finger protein PZF) (Penta Zf protein). | |||||
|
ANKS3_MOUSE
|
||||||
| NC score | 0.005915 (rank : 49) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CZK6, Q6ZPF6, Q80X46 | Gene names | Anks3, Kiaa1977 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 3. | |||||
|
PRP4B_HUMAN
|
||||||
| NC score | 0.004343 (rank : 50) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
CADH3_HUMAN
|
||||||
| NC score | 0.003476 (rank : 51) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22223 | Gene names | CDH3, CDHP | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-3 precursor (Placental-cadherin) (P-cadherin). | |||||