Please be patient as the page loads
|
HYES_MOUSE
|
||||||
| SwissProt Accessions | P34914, Q8CGV0 | Gene names | Ephx2, Eph2 | |||
|
Domain Architecture |

|
|||||
| Description | Epoxide hydrolase 2 (EC 3.3.2.10) (Soluble epoxide hydrolase) (SEH) (Epoxide hydratase) (Cytosolic epoxide hydrolase) (CEH). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HYES_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.991298 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P34913, Q16764, Q9HBJ1, Q9HBJ2 | Gene names | EPHX2 | |||
|
Domain Architecture |

|
|||||
| Description | Epoxide hydrolase 2 (EC 3.3.2.10) (Soluble epoxide hydrolase) (SEH) (Epoxide hydratase) (Cytosolic epoxide hydrolase) (CEH). | |||||
|
HYES_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P34914, Q8CGV0 | Gene names | Ephx2, Eph2 | |||
|
Domain Architecture |

|
|||||
| Description | Epoxide hydrolase 2 (EC 3.3.2.10) (Soluble epoxide hydrolase) (SEH) (Epoxide hydratase) (Cytosolic epoxide hydrolase) (CEH). | |||||
|
ABHD4_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 3) | NC score | 0.296124 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VD66 | Gene names | Abhd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 4 (EC 3.1.1.-) (Lyso-N- acylphosphatidylethanolamine lipase) (Alpha/beta-hydrolase 4). | |||||
|
ABHD4_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 4) | NC score | 0.291771 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TB40, Q9H9E0 | Gene names | ABHD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 4 (EC 3.1.1.-) (Lyso-N- acylphosphatidylethanolamine lipase) (Alpha/beta-hydrolase 4). | |||||
|
HYEP_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 5) | NC score | 0.208858 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D379 | Gene names | Ephx1 | |||
|
Domain Architecture |
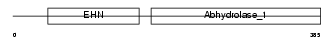
|
|||||
| Description | Epoxide hydrolase 1 (EC 3.3.2.9) (Microsomal epoxide hydrolase) (Epoxide hydratase). | |||||
|
SERHL_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 6) | NC score | 0.252718 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H4I8, Q5H973, Q9BR29, Q9BR30, Q9Y3I9 | Gene names | SERHL | |||
|
Domain Architecture |
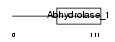
|
|||||
| Description | Serine hydrolase-like protein (EC 3.1.-.-). | |||||
|
ABHD5_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 7) | NC score | 0.258744 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WTS1, Q9Y369 | Gene names | ABHD5, NCIE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 5. | |||||
|
SEHL2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 8) | NC score | 0.249672 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQF3, Q5JZ95, Q9UH21 | Gene names | SERHL2 | |||
|
Domain Architecture |
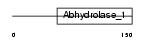
|
|||||
| Description | Serine hydrolase-like protein 2 (EC 3.1.-.-). | |||||
|
HYEP_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 9) | NC score | 0.187006 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P07099, Q5VTJ6, Q9NP75, Q9NPE7, Q9NQU6, Q9NQU7, Q9NQU8, Q9NQU9, Q9NQV0, Q9NQV1, Q9NQV2 | Gene names | EPHX1, EPHX, EPOX | |||
|
Domain Architecture |
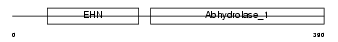
|
|||||
| Description | Epoxide hydrolase 1 (EC 3.3.2.9) (Microsomal epoxide hydrolase) (Epoxide hydratase). | |||||
|
ABHD5_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 10) | NC score | 0.251572 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DBL9, Q9CTY3 | Gene names | Abhd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 5 (Lipid droplet-binding protein CGI-58). | |||||
|
PPME1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 11) | NC score | 0.164190 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BVQ5, Q3U392, Q3UJX8, Q3UKE0, Q8K311, Q91YR8, Q9CSP7 | Gene names | Ppme1, Pme1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein phosphatase methylesterase 1 (EC 3.1.1.-) (PME-1). | |||||
|
CN029_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 12) | NC score | 0.105109 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z5M8, Q7Z5M6, Q7Z5M7, Q8N4D2 | Gene names | C14orf29 | |||
|
Domain Architecture |
|
|||||
| Description | Protein C14orf29. | |||||
|
SERHL_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 13) | NC score | 0.216455 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EPB5, Q9DCZ8 | Gene names | Serhl | |||
|
Domain Architecture |
|
|||||
| Description | Serine hydrolase-like protein (EC 3.1.-.-) (SHL). | |||||
|
PPME1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.108509 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y570, Q9NVT5, Q9UI18 | Gene names | PPME1, PME1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein phosphatase methylesterase 1 (EC 3.1.1.-) (PME-1). | |||||
|
AB14B_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.138120 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96IU4, Q86VK8, Q8N8W5 | Gene names | ABHD14B, CIB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 14B (CCG1-interacting factor B). | |||||
|
MET7B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.059129 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DD20, Q78ID2 | Gene names | Mettl7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyltransferase-like protein 7B precursor (EC 2.1.1.-). | |||||
|
AB14B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.112895 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCR7 | Gene names | Abhd14b, Cib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 14B (CCG1-interacting factor B). | |||||
|
ELMO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.026732 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92556, Q29R79, Q96PB0, Q9H0I1 | Gene names | ELMO1, KIAA0281 | |||
|
Domain Architecture |

|
|||||
| Description | Engulfment and cell motility protein 1 (CED-12 homolog). | |||||
|
ELMO1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.026748 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BPU7, Q8BSY9, Q8K2C5, Q91ZU3 | Gene names | Elmo1 | |||
|
Domain Architecture |

|
|||||
| Description | Engulfment and cell motility protein 1 (CED-12 homolog). | |||||
|
KIT_HUMAN
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.007341 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 953 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10721, Q9UM99 | Gene names | KIT | |||
|
Domain Architecture |

|
|||||
| Description | Mast/stem cell growth factor receptor precursor (EC 2.7.10.1) (SCFR) (Proto-oncogene tyrosine-protein kinase Kit) (c-kit) (CD117 antigen). | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.012019 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
XTP3B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.021831 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96DZ1, O95901, Q6UWN7, Q9NUY7, Q9UQL4 | Gene names | C2orf30, XTP3TPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XTP3-transactivated gene B protein precursor. | |||||
|
CT135_HUMAN
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.025015 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H3Z7 | Gene names | C20orf135 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf135. | |||||
|
KIT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.003819 (rank : 25) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05532, Q61415, Q61416, Q61417, Q6LEE9 | Gene names | Kit, Sl | |||
|
Domain Architecture |

|
|||||
| Description | Mast/stem cell growth factor receptor precursor (EC 2.7.10.1) (SCFR) (Proto-oncogene tyrosine-protein kinase Kit) (c-kit) (CD117 antigen). | |||||
|
BPHL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.060072 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R164, Q8BVH2, Q8R589, Q9DCC6 | Gene names | Bphl | |||
|
Domain Architecture |
|
|||||
| Description | Valacyclovir hydrolase precursor (EC 3.1.-.-) (VACVase) (Biphenyl hydrolase-like protein). | |||||
|
HYES_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P34914, Q8CGV0 | Gene names | Ephx2, Eph2 | |||
|
Domain Architecture |

|
|||||
| Description | Epoxide hydrolase 2 (EC 3.3.2.10) (Soluble epoxide hydrolase) (SEH) (Epoxide hydratase) (Cytosolic epoxide hydrolase) (CEH). | |||||
|
HYES_HUMAN
|
||||||
| NC score | 0.991298 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P34913, Q16764, Q9HBJ1, Q9HBJ2 | Gene names | EPHX2 | |||
|
Domain Architecture |

|
|||||
| Description | Epoxide hydrolase 2 (EC 3.3.2.10) (Soluble epoxide hydrolase) (SEH) (Epoxide hydratase) (Cytosolic epoxide hydrolase) (CEH). | |||||
|
ABHD4_MOUSE
|
||||||
| NC score | 0.296124 (rank : 3) | θ value | 6.43352e-06 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VD66 | Gene names | Abhd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 4 (EC 3.1.1.-) (Lyso-N- acylphosphatidylethanolamine lipase) (Alpha/beta-hydrolase 4). | |||||
|
ABHD4_HUMAN
|
||||||
| NC score | 0.291771 (rank : 4) | θ value | 2.44474e-05 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TB40, Q9H9E0 | Gene names | ABHD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 4 (EC 3.1.1.-) (Lyso-N- acylphosphatidylethanolamine lipase) (Alpha/beta-hydrolase 4). | |||||
|
ABHD5_HUMAN
|
||||||
| NC score | 0.258744 (rank : 5) | θ value | 0.00665767 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WTS1, Q9Y369 | Gene names | ABHD5, NCIE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 5. | |||||
|
SERHL_HUMAN
|
||||||
| NC score | 0.252718 (rank : 6) | θ value | 0.00134147 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H4I8, Q5H973, Q9BR29, Q9BR30, Q9Y3I9 | Gene names | SERHL | |||
|
Domain Architecture |

|
|||||
| Description | Serine hydrolase-like protein (EC 3.1.-.-). | |||||
|
ABHD5_MOUSE
|
||||||
| NC score | 0.251572 (rank : 7) | θ value | 0.0252991 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DBL9, Q9CTY3 | Gene names | Abhd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 5 (Lipid droplet-binding protein CGI-58). | |||||
|
SEHL2_HUMAN
|
||||||
| NC score | 0.249672 (rank : 8) | θ value | 0.00869519 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQF3, Q5JZ95, Q9UH21 | Gene names | SERHL2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine hydrolase-like protein 2 (EC 3.1.-.-). | |||||
|
SERHL_MOUSE
|
||||||
| NC score | 0.216455 (rank : 9) | θ value | 0.0736092 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EPB5, Q9DCZ8 | Gene names | Serhl | |||
|
Domain Architecture |

|
|||||
| Description | Serine hydrolase-like protein (EC 3.1.-.-) (SHL). | |||||
|
HYEP_MOUSE
|
||||||
| NC score | 0.208858 (rank : 10) | θ value | 0.000461057 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D379 | Gene names | Ephx1 | |||
|
Domain Architecture |

|
|||||
| Description | Epoxide hydrolase 1 (EC 3.3.2.9) (Microsomal epoxide hydrolase) (Epoxide hydratase). | |||||
|
HYEP_HUMAN
|
||||||
| NC score | 0.187006 (rank : 11) | θ value | 0.0148317 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P07099, Q5VTJ6, Q9NP75, Q9NPE7, Q9NQU6, Q9NQU7, Q9NQU8, Q9NQU9, Q9NQV0, Q9NQV1, Q9NQV2 | Gene names | EPHX1, EPHX, EPOX | |||
|
Domain Architecture |

|
|||||
| Description | Epoxide hydrolase 1 (EC 3.3.2.9) (Microsomal epoxide hydrolase) (Epoxide hydratase). | |||||
|
PPME1_MOUSE
|
||||||
| NC score | 0.164190 (rank : 12) | θ value | 0.0252991 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BVQ5, Q3U392, Q3UJX8, Q3UKE0, Q8K311, Q91YR8, Q9CSP7 | Gene names | Ppme1, Pme1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase methylesterase 1 (EC 3.1.1.-) (PME-1). | |||||
|
AB14B_HUMAN
|
||||||
| NC score | 0.138120 (rank : 13) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96IU4, Q86VK8, Q8N8W5 | Gene names | ABHD14B, CIB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 14B (CCG1-interacting factor B). | |||||
|
AB14B_MOUSE
|
||||||
| NC score | 0.112895 (rank : 14) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCR7 | Gene names | Abhd14b, Cib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 14B (CCG1-interacting factor B). | |||||
|
PPME1_HUMAN
|
||||||
| NC score | 0.108509 (rank : 15) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y570, Q9NVT5, Q9UI18 | Gene names | PPME1, PME1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase methylesterase 1 (EC 3.1.1.-) (PME-1). | |||||
|
CN029_HUMAN
|
||||||
| NC score | 0.105109 (rank : 16) | θ value | 0.0563607 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z5M8, Q7Z5M6, Q7Z5M7, Q8N4D2 | Gene names | C14orf29 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C14orf29. | |||||
|
BPHL_MOUSE
|
||||||
| NC score | 0.060072 (rank : 17) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R164, Q8BVH2, Q8R589, Q9DCC6 | Gene names | Bphl | |||
|
Domain Architecture |

|
|||||
| Description | Valacyclovir hydrolase precursor (EC 3.1.-.-) (VACVase) (Biphenyl hydrolase-like protein). | |||||
|
MET7B_MOUSE
|
||||||
| NC score | 0.059129 (rank : 18) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DD20, Q78ID2 | Gene names | Mettl7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyltransferase-like protein 7B precursor (EC 2.1.1.-). | |||||
|
ELMO1_MOUSE
|
||||||
| NC score | 0.026748 (rank : 19) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BPU7, Q8BSY9, Q8K2C5, Q91ZU3 | Gene names | Elmo1 | |||
|
Domain Architecture |

|
|||||
| Description | Engulfment and cell motility protein 1 (CED-12 homolog). | |||||
|
ELMO1_HUMAN
|
||||||
| NC score | 0.026732 (rank : 20) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92556, Q29R79, Q96PB0, Q9H0I1 | Gene names | ELMO1, KIAA0281 | |||
|
Domain Architecture |

|
|||||
| Description | Engulfment and cell motility protein 1 (CED-12 homolog). | |||||
|
CT135_HUMAN
|
||||||
| NC score | 0.025015 (rank : 21) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H3Z7 | Gene names | C20orf135 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf135. | |||||
|
XTP3B_HUMAN
|
||||||
| NC score | 0.021831 (rank : 22) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96DZ1, O95901, Q6UWN7, Q9NUY7, Q9UQL4 | Gene names | C2orf30, XTP3TPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XTP3-transactivated gene B protein precursor. | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.012019 (rank : 23) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
KIT_HUMAN
|
||||||
| NC score | 0.007341 (rank : 24) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 953 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10721, Q9UM99 | Gene names | KIT | |||
|
Domain Architecture |

|
|||||
| Description | Mast/stem cell growth factor receptor precursor (EC 2.7.10.1) (SCFR) (Proto-oncogene tyrosine-protein kinase Kit) (c-kit) (CD117 antigen). | |||||
|
KIT_MOUSE
|
||||||
| NC score | 0.003819 (rank : 25) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05532, Q61415, Q61416, Q61417, Q6LEE9 | Gene names | Kit, Sl | |||
|
Domain Architecture |

|
|||||
| Description | Mast/stem cell growth factor receptor precursor (EC 2.7.10.1) (SCFR) (Proto-oncogene tyrosine-protein kinase Kit) (c-kit) (CD117 antigen). | |||||