Please be patient as the page loads
|
ABHD4_HUMAN
|
||||||
| SwissProt Accessions | Q8TB40, Q9H9E0 | Gene names | ABHD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 4 (EC 3.1.1.-) (Lyso-N- acylphosphatidylethanolamine lipase) (Alpha/beta-hydrolase 4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ABHD4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TB40, Q9H9E0 | Gene names | ABHD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 4 (EC 3.1.1.-) (Lyso-N- acylphosphatidylethanolamine lipase) (Alpha/beta-hydrolase 4). | |||||
|
ABHD4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998034 (rank : 2) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VD66 | Gene names | Abhd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 4 (EC 3.1.1.-) (Lyso-N- acylphosphatidylethanolamine lipase) (Alpha/beta-hydrolase 4). | |||||
|
ABHD5_HUMAN
|
||||||
| θ value | 8.71062e-112 (rank : 3) | NC score | 0.977605 (rank : 3) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WTS1, Q9Y369 | Gene names | ABHD5, NCIE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 5. | |||||
|
ABHD5_MOUSE
|
||||||
| θ value | 8.15288e-110 (rank : 4) | NC score | 0.973403 (rank : 4) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBL9, Q9CTY3 | Gene names | Abhd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 5 (Lipid droplet-binding protein CGI-58). | |||||
|
HYES_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 5) | NC score | 0.291771 (rank : 5) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P34914, Q8CGV0 | Gene names | Ephx2, Eph2 | |||
|
Domain Architecture |

|
|||||
| Description | Epoxide hydrolase 2 (EC 3.3.2.10) (Soluble epoxide hydrolase) (SEH) (Epoxide hydratase) (Cytosolic epoxide hydrolase) (CEH). | |||||
|
HYES_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 6) | NC score | 0.282745 (rank : 6) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P34913, Q16764, Q9HBJ1, Q9HBJ2 | Gene names | EPHX2 | |||
|
Domain Architecture |

|
|||||
| Description | Epoxide hydrolase 2 (EC 3.3.2.10) (Soluble epoxide hydrolase) (SEH) (Epoxide hydratase) (Cytosolic epoxide hydrolase) (CEH). | |||||
|
SERHL_MOUSE
|
||||||
| θ value | 0.279714 (rank : 7) | NC score | 0.144688 (rank : 7) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9EPB5, Q9DCZ8 | Gene names | Serhl | |||
|
Domain Architecture |
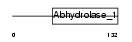
|
|||||
| Description | Serine hydrolase-like protein (EC 3.1.-.-) (SHL). | |||||
|
SFRP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 8) | NC score | 0.036149 (rank : 15) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96HF1, O14778, Q9HAP5 | Gene names | SFRP2, FRP2, SARP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 2 precursor (sFRP-2) (Secreted apoptosis-related protein 1) (SARP-1) (Frizzled-related protein 2) (FRP-2). | |||||
|
SFRP2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 9) | NC score | 0.036125 (rank : 16) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97299, O08862, O35297 | Gene names | Sfrp2, Sarp1, Sdf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 2 precursor (sFRP-2) (Secreted apoptosis-related protein 1) (SARP-1) (SDF5 protein). | |||||
|
BPHL_MOUSE
|
||||||
| θ value | 1.38821 (rank : 10) | NC score | 0.098686 (rank : 8) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R164, Q8BVH2, Q8R589, Q9DCC6 | Gene names | Bphl | |||
|
Domain Architecture |
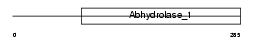
|
|||||
| Description | Valacyclovir hydrolase precursor (EC 3.1.-.-) (VACVase) (Biphenyl hydrolase-like protein). | |||||
|
LIPR2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 11) | NC score | 0.034125 (rank : 17) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54317 | Gene names | PNLIPRP2, PLRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic lipase-related protein 2 precursor (EC 3.1.1.3). | |||||
|
NFASC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 12) | NC score | 0.010487 (rank : 22) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94856, Q5T2F0, Q5T2F1, Q5T2F2, Q5T2F3, Q5T2F4, Q5T2F5, Q5T2F6, Q5T2F7, Q5T2F9, Q5T2G0, Q5W9F8, Q68DH3, Q6ZQV6, Q7Z3K1, Q96HT1, Q96K50 | Gene names | NFASC, KIAA0756 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofascin precursor. | |||||
|
TSYL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 13) | NC score | 0.018160 (rank : 19) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88852, Q3TKW0 | Gene names | Tspyl1, Tspyl | |||
|
Domain Architecture |
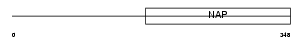
|
|||||
| Description | Testis-specific Y-encoded-like protein 1 (TSPY-like 1). | |||||
|
CT135_HUMAN
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.043407 (rank : 14) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H3Z7 | Gene names | C20orf135 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf135. | |||||
|
CT135_MOUSE
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.044933 (rank : 13) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80YU0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf135 homolog. | |||||
|
LIPR2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 16) | NC score | 0.017876 (rank : 20) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17892 | Gene names | Pnliprp2, Plrp2 | |||
|
Domain Architecture |
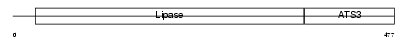
|
|||||
| Description | Pancreatic lipase-related protein 2 precursor (EC 3.1.1.3) (Cytotoxic T-lymphocyte lipase). | |||||
|
BAT5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.025436 (rank : 18) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95870, Q5SRR1, Q5SRR2, Q8WYH0, Q9NW33 | Gene names | BAT5, G5, NG26 | |||
|
Domain Architecture |

|
|||||
| Description | Protein BAT5 (HLA-B-associated transcript 5) (Protein G5). | |||||
|
KDEL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 18) | NC score | 0.010635 (rank : 21) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z4H8, Q6UWW2, Q6ZUM9, Q8N7L8, Q8NE24 | Gene names | KDELC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KDEL motif-containing protein 2 precursor. | |||||
|
TEP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 19) | NC score | 0.007935 (rank : 23) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99973 | Gene names | TEP1, TLP1, TP1 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase protein component 1 (Telomerase-associated protein 1) (Telomerase protein 1) (p240) (p80 telomerase homolog). | |||||
|
BPHL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 20) | NC score | 0.092094 (rank : 10) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86WA6, Q00306, Q13855 | Gene names | BPHL | |||
|
Domain Architecture |
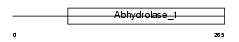
|
|||||
| Description | Valacyclovir hydrolase precursor (EC 3.1.-.-) (VACVase) (Biphenyl hydrolase-like protein) (Biphenyl hydrolase-related protein) (Bph-rp) (Breast epithelial mucin-associated antigen) (MCNAA). | |||||
|
MGLL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 21) | NC score | 0.057928 (rank : 12) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99685, Q96AA5 | Gene names | MGLL | |||
|
Domain Architecture |
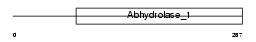
|
|||||
| Description | Monoglyceride lipase (EC 3.1.1.23) (MGL) (HU-K5) (Lysophospholipase homolog) (Lysophospholipase-like). | |||||
|
SEHL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 22) | NC score | 0.092939 (rank : 9) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NQF3, Q5JZ95, Q9UH21 | Gene names | SERHL2 | |||
|
Domain Architecture |
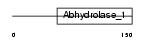
|
|||||
| Description | Serine hydrolase-like protein 2 (EC 3.1.-.-). | |||||
|
SERHL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.089058 (rank : 11) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H4I8, Q5H973, Q9BR29, Q9BR30, Q9Y3I9 | Gene names | SERHL | |||
|
Domain Architecture |
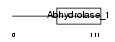
|
|||||
| Description | Serine hydrolase-like protein (EC 3.1.-.-). | |||||
|
ABHD4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TB40, Q9H9E0 | Gene names | ABHD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 4 (EC 3.1.1.-) (Lyso-N- acylphosphatidylethanolamine lipase) (Alpha/beta-hydrolase 4). | |||||
|
ABHD4_MOUSE
|
||||||
| NC score | 0.998034 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VD66 | Gene names | Abhd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 4 (EC 3.1.1.-) (Lyso-N- acylphosphatidylethanolamine lipase) (Alpha/beta-hydrolase 4). | |||||
|
ABHD5_HUMAN
|
||||||
| NC score | 0.977605 (rank : 3) | θ value | 8.71062e-112 (rank : 3) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WTS1, Q9Y369 | Gene names | ABHD5, NCIE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 5. | |||||
|
ABHD5_MOUSE
|
||||||
| NC score | 0.973403 (rank : 4) | θ value | 8.15288e-110 (rank : 4) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBL9, Q9CTY3 | Gene names | Abhd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abhydrolase domain-containing protein 5 (Lipid droplet-binding protein CGI-58). | |||||
|
HYES_MOUSE
|
||||||
| NC score | 0.291771 (rank : 5) | θ value | 2.44474e-05 (rank : 5) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P34914, Q8CGV0 | Gene names | Ephx2, Eph2 | |||
|
Domain Architecture |

|
|||||
| Description | Epoxide hydrolase 2 (EC 3.3.2.10) (Soluble epoxide hydrolase) (SEH) (Epoxide hydratase) (Cytosolic epoxide hydrolase) (CEH). | |||||
|
HYES_HUMAN
|
||||||
| NC score | 0.282745 (rank : 6) | θ value | 0.000158464 (rank : 6) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P34913, Q16764, Q9HBJ1, Q9HBJ2 | Gene names | EPHX2 | |||
|
Domain Architecture |

|
|||||
| Description | Epoxide hydrolase 2 (EC 3.3.2.10) (Soluble epoxide hydrolase) (SEH) (Epoxide hydratase) (Cytosolic epoxide hydrolase) (CEH). | |||||
|
SERHL_MOUSE
|
||||||
| NC score | 0.144688 (rank : 7) | θ value | 0.279714 (rank : 7) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9EPB5, Q9DCZ8 | Gene names | Serhl | |||
|
Domain Architecture |

|
|||||
| Description | Serine hydrolase-like protein (EC 3.1.-.-) (SHL). | |||||
|
BPHL_MOUSE
|
||||||
| NC score | 0.098686 (rank : 8) | θ value | 1.38821 (rank : 10) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R164, Q8BVH2, Q8R589, Q9DCC6 | Gene names | Bphl | |||
|
Domain Architecture |

|
|||||
| Description | Valacyclovir hydrolase precursor (EC 3.1.-.-) (VACVase) (Biphenyl hydrolase-like protein). | |||||
|
SEHL2_HUMAN
|
||||||
| NC score | 0.092939 (rank : 9) | θ value | θ > 10 (rank : 22) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NQF3, Q5JZ95, Q9UH21 | Gene names | SERHL2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine hydrolase-like protein 2 (EC 3.1.-.-). | |||||
|
BPHL_HUMAN
|
||||||
| NC score | 0.092094 (rank : 10) | θ value | θ > 10 (rank : 20) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86WA6, Q00306, Q13855 | Gene names | BPHL | |||
|
Domain Architecture |

|
|||||
| Description | Valacyclovir hydrolase precursor (EC 3.1.-.-) (VACVase) (Biphenyl hydrolase-like protein) (Biphenyl hydrolase-related protein) (Bph-rp) (Breast epithelial mucin-associated antigen) (MCNAA). | |||||
|
SERHL_HUMAN
|
||||||
| NC score | 0.089058 (rank : 11) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H4I8, Q5H973, Q9BR29, Q9BR30, Q9Y3I9 | Gene names | SERHL | |||
|
Domain Architecture |

|
|||||
| Description | Serine hydrolase-like protein (EC 3.1.-.-). | |||||
|
MGLL_HUMAN
|
||||||
| NC score | 0.057928 (rank : 12) | θ value | θ > 10 (rank : 21) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99685, Q96AA5 | Gene names | MGLL | |||
|
Domain Architecture |

|
|||||
| Description | Monoglyceride lipase (EC 3.1.1.23) (MGL) (HU-K5) (Lysophospholipase homolog) (Lysophospholipase-like). | |||||
|
CT135_MOUSE
|
||||||
| NC score | 0.044933 (rank : 13) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80YU0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf135 homolog. | |||||
|
CT135_HUMAN
|
||||||
| NC score | 0.043407 (rank : 14) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H3Z7 | Gene names | C20orf135 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf135. | |||||
|
SFRP2_HUMAN
|
||||||
| NC score | 0.036149 (rank : 15) | θ value | 1.06291 (rank : 8) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96HF1, O14778, Q9HAP5 | Gene names | SFRP2, FRP2, SARP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 2 precursor (sFRP-2) (Secreted apoptosis-related protein 1) (SARP-1) (Frizzled-related protein 2) (FRP-2). | |||||
|
SFRP2_MOUSE
|
||||||
| NC score | 0.036125 (rank : 16) | θ value | 1.06291 (rank : 9) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97299, O08862, O35297 | Gene names | Sfrp2, Sarp1, Sdf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted frizzled-related protein 2 precursor (sFRP-2) (Secreted apoptosis-related protein 1) (SARP-1) (SDF5 protein). | |||||
|
LIPR2_HUMAN
|
||||||
| NC score | 0.034125 (rank : 17) | θ value | 1.38821 (rank : 11) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54317 | Gene names | PNLIPRP2, PLRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic lipase-related protein 2 precursor (EC 3.1.1.3). | |||||
|
BAT5_HUMAN
|
||||||
| NC score | 0.025436 (rank : 18) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95870, Q5SRR1, Q5SRR2, Q8WYH0, Q9NW33 | Gene names | BAT5, G5, NG26 | |||
|
Domain Architecture |

|
|||||
| Description | Protein BAT5 (HLA-B-associated transcript 5) (Protein G5). | |||||
|
TSYL1_MOUSE
|
||||||
| NC score | 0.018160 (rank : 19) | θ value | 2.36792 (rank : 13) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88852, Q3TKW0 | Gene names | Tspyl1, Tspyl | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific Y-encoded-like protein 1 (TSPY-like 1). | |||||
|
LIPR2_MOUSE
|
||||||
| NC score | 0.017876 (rank : 20) | θ value | 6.88961 (rank : 16) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17892 | Gene names | Pnliprp2, Plrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic lipase-related protein 2 precursor (EC 3.1.1.3) (Cytotoxic T-lymphocyte lipase). | |||||
|
KDEL2_HUMAN
|
||||||
| NC score | 0.010635 (rank : 21) | θ value | 8.99809 (rank : 18) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z4H8, Q6UWW2, Q6ZUM9, Q8N7L8, Q8NE24 | Gene names | KDELC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KDEL motif-containing protein 2 precursor. | |||||
|
NFASC_HUMAN
|
||||||
| NC score | 0.010487 (rank : 22) | θ value | 2.36792 (rank : 12) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94856, Q5T2F0, Q5T2F1, Q5T2F2, Q5T2F3, Q5T2F4, Q5T2F5, Q5T2F6, Q5T2F7, Q5T2F9, Q5T2G0, Q5W9F8, Q68DH3, Q6ZQV6, Q7Z3K1, Q96HT1, Q96K50 | Gene names | NFASC, KIAA0756 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofascin precursor. | |||||
|
TEP1_HUMAN
|
||||||
| NC score | 0.007935 (rank : 23) | θ value | 8.99809 (rank : 19) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99973 | Gene names | TEP1, TLP1, TP1 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase protein component 1 (Telomerase-associated protein 1) (Telomerase protein 1) (p240) (p80 telomerase homolog). | |||||