Please be patient as the page loads
|
HSPB1_MOUSE
|
||||||
| SwissProt Accessions | P14602 | Gene names | Hspb1, Hsp25, Hsp27 | |||
|
Domain Architecture |
|
|||||
| Description | Heat-shock protein beta-1 (HspB1) (Heat shock 27 kDa protein) (HSP 27) (Growth-related 25 kDa protein) (P25) (HSP25). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HSPB1_MOUSE
|
||||||
| θ value | 8.69046e-120 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P14602 | Gene names | Hspb1, Hsp25, Hsp27 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-1 (HspB1) (Heat shock 27 kDa protein) (HSP 27) (Growth-related 25 kDa protein) (P25) (HSP25). | |||||
|
HSPB1_HUMAN
|
||||||
| θ value | 7.64859e-100 (rank : 2) | NC score | 0.998002 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P04792, Q6FI47, Q9UC31 | Gene names | HSPB1, HSP27 | |||
|
Domain Architecture |
|
|||||
| Description | Heat-shock protein beta-1 (HspB1) (Heat shock 27 kDa protein) (HSP 27) (Stress-responsive protein 27) (SRP27) (Estrogen-regulated 24 kDa protein) (28 kDa heat shock protein). | |||||
|
CRYAB_MOUSE
|
||||||
| θ value | 1.2049e-28 (rank : 3) | NC score | 0.913772 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23927, Q64325 | Gene names | Cryab, Crya2 | |||
|
Domain Architecture |
|
|||||
| Description | Alpha crystallin B chain (Alpha(B)-crystallin) (P23). | |||||
|
CRYAB_HUMAN
|
||||||
| θ value | 3.50572e-28 (rank : 4) | NC score | 0.912472 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P02511, O43416 | Gene names | CRYAB, CRYA2 | |||
|
Domain Architecture |
|
|||||
| Description | Alpha crystallin B chain (Alpha(B)-crystallin) (Rosenthal fiber component) (Heat-shock protein beta-5) (HspB5) (NY-REN-27 antigen). | |||||
|
HSPB8_MOUSE
|
||||||
| θ value | 2.12685e-25 (rank : 5) | NC score | 0.887426 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JK92 | Gene names | Hspb8, Cryac, Hsp22 | |||
|
Domain Architecture |
|
|||||
| Description | Heat-shock protein beta-8 (HspB8) (Alpha crystallin C chain) (Small stress protein-like protein HSP22). | |||||
|
HSPB8_HUMAN
|
||||||
| θ value | 2.77775e-25 (rank : 6) | NC score | 0.886424 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UJY1, Q6FIH3, Q9UKS3 | Gene names | HSPB8, CRYAC, E2IG1, HSP22 | |||
|
Domain Architecture |
|
|||||
| Description | Heat-shock protein beta-8 (HspB8) (Alpha crystallin C chain) (Small stress protein-like protein HSP22) (E2-induced gene 1 protein) (Protein kinase H11). | |||||
|
CRYAA_HUMAN
|
||||||
| θ value | 9.87957e-23 (rank : 7) | NC score | 0.889126 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P02489 | Gene names | CRYAA, CRYA1, HSPB4 | |||
|
Domain Architecture |
|
|||||
| Description | Alpha crystallin A chain (Heat-shock protein beta-4) (HspB4) [Contains: Alpha crystallin A chain, short form]. | |||||
|
HSPB6_MOUSE
|
||||||
| θ value | 1.42661e-21 (rank : 8) | NC score | 0.903121 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5EBG6 | Gene names | Hspb6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat-shock protein beta-6 (HspB6). | |||||
|
CRYAA_MOUSE
|
||||||
| θ value | 3.51386e-20 (rank : 9) | NC score | 0.876778 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P24622, P02490, P02496, P82532, Q61444, Q6LEL4 | Gene names | Cryaa, Crya1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha crystallin A chain [Contains: Alpha crystallin A chain, short form]. | |||||
|
HSPB2_HUMAN
|
||||||
| θ value | 5.99374e-20 (rank : 10) | NC score | 0.876615 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q16082, Q6I9U7 | Gene names | HSPB2 | |||
|
Domain Architecture |
|
|||||
| Description | Heat-shock protein beta-2 (HspB2) (DMPK-binding protein) (MKBP). | |||||
|
HSPB6_HUMAN
|
||||||
| θ value | 7.82807e-20 (rank : 11) | NC score | 0.901395 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14558, O14551, Q6NVI3, Q96MG9 | Gene names | HSPB6 | |||
|
Domain Architecture |
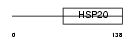
|
|||||
| Description | Heat-shock protein beta-6 (HspB6) (Heat-shock 20 kDa-like protein p20). | |||||
|
HSPB2_MOUSE
|
||||||
| θ value | 1.74391e-19 (rank : 12) | NC score | 0.876719 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PR8, Q9CZC2 | Gene names | Hspb2 | |||
|
Domain Architecture |
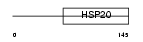
|
|||||
| Description | Heat-shock protein beta-2 (HspB2). | |||||
|
HSPB3_MOUSE
|
||||||
| θ value | 7.84624e-12 (rank : 13) | NC score | 0.785453 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZ57, Q569N3 | Gene names | Hspb3 | |||
|
Domain Architecture |
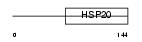
|
|||||
| Description | Heat-shock protein beta-3 (HspB3). | |||||
|
HSPB3_HUMAN
|
||||||
| θ value | 1.02475e-11 (rank : 14) | NC score | 0.781390 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12988 | Gene names | HSPB3, HSPL27 | |||
|
Domain Architecture |
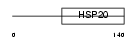
|
|||||
| Description | Heat-shock protein beta-3 (HspB3) (Heat shock 17 kDa protein). | |||||
|
HSPB7_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 15) | NC score | 0.662195 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UBY9, Q9NU17 | Gene names | HSPB7, CVHSP | |||
|
Domain Architecture |
|
|||||
| Description | Heat-shock protein beta-7 (HspB7) (Cardiovascular heat shock protein) (cvHsp). | |||||
|
HSPB7_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 16) | NC score | 0.646061 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35385, Q8CDI0, Q9QUS2 | Gene names | Hspb7, Cvhsp, Hsp25-2 | |||
|
Domain Architecture |
|
|||||
| Description | Heat-shock protein beta-7 (HspB7) (Cardiovascular heat shock protein) (cvHsp) (Heat shock protein 25 kDa 2) (Protein p19/6.8). | |||||
|
HSPB9_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 17) | NC score | 0.482201 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9DAM3 | Gene names | Hspb9 | |||
|
Domain Architecture |
|
|||||
| Description | Heat-shock protein beta-9 (HspB9). | |||||
|
ODFP_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 18) | NC score | 0.220523 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14990 | Gene names | ODF1, ODFP | |||
|
Domain Architecture |

|
|||||
| Description | Outer dense fiber protein. | |||||
|
ODFP_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 19) | NC score | 0.214751 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61999 | Gene names | Odf1, Odfp | |||
|
Domain Architecture |
|
|||||
| Description | Outer dense fiber protein. | |||||
|
HSPB9_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 20) | NC score | 0.412327 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BQS6, Q52LB4 | Gene names | HSPB9 | |||
|
Domain Architecture |
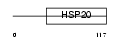
|
|||||
| Description | Heat-shock protein beta-9 (HspB9). | |||||
|
AFAD_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.023428 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
MMP17_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.008829 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULZ9, Q14850 | Gene names | MMP17, MT4MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
GAW2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.024659 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NRZ4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-W_3q26.32 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein (Gag P15); Capsid protein (Core shell protein) (Gag P30)]. | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.007367 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
CBPN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.009885 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJN5, Q91WM9 | Gene names | Cpn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase N catalytic chain precursor (EC 3.4.17.3) (CPN) (Carboxypeptidase N polypeptide 1) (Carboxypeptidase N small subunit). | |||||
|
WBS14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.018648 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
AKAP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.008421 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
HSPB1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 8.69046e-120 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P14602 | Gene names | Hspb1, Hsp25, Hsp27 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-1 (HspB1) (Heat shock 27 kDa protein) (HSP 27) (Growth-related 25 kDa protein) (P25) (HSP25). | |||||
|
HSPB1_HUMAN
|
||||||
| NC score | 0.998002 (rank : 2) | θ value | 7.64859e-100 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P04792, Q6FI47, Q9UC31 | Gene names | HSPB1, HSP27 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-1 (HspB1) (Heat shock 27 kDa protein) (HSP 27) (Stress-responsive protein 27) (SRP27) (Estrogen-regulated 24 kDa protein) (28 kDa heat shock protein). | |||||
|
CRYAB_MOUSE
|
||||||
| NC score | 0.913772 (rank : 3) | θ value | 1.2049e-28 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23927, Q64325 | Gene names | Cryab, Crya2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha crystallin B chain (Alpha(B)-crystallin) (P23). | |||||
|
CRYAB_HUMAN
|
||||||
| NC score | 0.912472 (rank : 4) | θ value | 3.50572e-28 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P02511, O43416 | Gene names | CRYAB, CRYA2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha crystallin B chain (Alpha(B)-crystallin) (Rosenthal fiber component) (Heat-shock protein beta-5) (HspB5) (NY-REN-27 antigen). | |||||
|
HSPB6_MOUSE
|
||||||
| NC score | 0.903121 (rank : 5) | θ value | 1.42661e-21 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5EBG6 | Gene names | Hspb6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat-shock protein beta-6 (HspB6). | |||||
|
HSPB6_HUMAN
|
||||||
| NC score | 0.901395 (rank : 6) | θ value | 7.82807e-20 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14558, O14551, Q6NVI3, Q96MG9 | Gene names | HSPB6 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-6 (HspB6) (Heat-shock 20 kDa-like protein p20). | |||||
|
CRYAA_HUMAN
|
||||||
| NC score | 0.889126 (rank : 7) | θ value | 9.87957e-23 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P02489 | Gene names | CRYAA, CRYA1, HSPB4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha crystallin A chain (Heat-shock protein beta-4) (HspB4) [Contains: Alpha crystallin A chain, short form]. | |||||
|
HSPB8_MOUSE
|
||||||
| NC score | 0.887426 (rank : 8) | θ value | 2.12685e-25 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JK92 | Gene names | Hspb8, Cryac, Hsp22 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-8 (HspB8) (Alpha crystallin C chain) (Small stress protein-like protein HSP22). | |||||
|
HSPB8_HUMAN
|
||||||
| NC score | 0.886424 (rank : 9) | θ value | 2.77775e-25 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UJY1, Q6FIH3, Q9UKS3 | Gene names | HSPB8, CRYAC, E2IG1, HSP22 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-8 (HspB8) (Alpha crystallin C chain) (Small stress protein-like protein HSP22) (E2-induced gene 1 protein) (Protein kinase H11). | |||||
|
CRYAA_MOUSE
|
||||||
| NC score | 0.876778 (rank : 10) | θ value | 3.51386e-20 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P24622, P02490, P02496, P82532, Q61444, Q6LEL4 | Gene names | Cryaa, Crya1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha crystallin A chain [Contains: Alpha crystallin A chain, short form]. | |||||
|
HSPB2_MOUSE
|
||||||
| NC score | 0.876719 (rank : 11) | θ value | 1.74391e-19 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q99PR8, Q9CZC2 | Gene names | Hspb2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-2 (HspB2). | |||||
|
HSPB2_HUMAN
|
||||||
| NC score | 0.876615 (rank : 12) | θ value | 5.99374e-20 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q16082, Q6I9U7 | Gene names | HSPB2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-2 (HspB2) (DMPK-binding protein) (MKBP). | |||||
|
HSPB3_MOUSE
|
||||||
| NC score | 0.785453 (rank : 13) | θ value | 7.84624e-12 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZ57, Q569N3 | Gene names | Hspb3 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-3 (HspB3). | |||||
|
HSPB3_HUMAN
|
||||||
| NC score | 0.781390 (rank : 14) | θ value | 1.02475e-11 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12988 | Gene names | HSPB3, HSPL27 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-3 (HspB3) (Heat shock 17 kDa protein). | |||||
|
HSPB7_HUMAN
|
||||||
| NC score | 0.662195 (rank : 15) | θ value | 2.61198e-07 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UBY9, Q9NU17 | Gene names | HSPB7, CVHSP | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-7 (HspB7) (Cardiovascular heat shock protein) (cvHsp). | |||||
|
HSPB7_MOUSE
|
||||||
| NC score | 0.646061 (rank : 16) | θ value | 2.21117e-06 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35385, Q8CDI0, Q9QUS2 | Gene names | Hspb7, Cvhsp, Hsp25-2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-7 (HspB7) (Cardiovascular heat shock protein) (cvHsp) (Heat shock protein 25 kDa 2) (Protein p19/6.8). | |||||
|
HSPB9_MOUSE
|
||||||
| NC score | 0.482201 (rank : 17) | θ value | 0.00035302 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9DAM3 | Gene names | Hspb9 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-9 (HspB9). | |||||
|
HSPB9_HUMAN
|
||||||
| NC score | 0.412327 (rank : 18) | θ value | 0.0252991 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BQS6, Q52LB4 | Gene names | HSPB9 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-9 (HspB9). | |||||
|
ODFP_HUMAN
|
||||||
| NC score | 0.220523 (rank : 19) | θ value | 0.0148317 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14990 | Gene names | ODF1, ODFP | |||
|
Domain Architecture |

|
|||||
| Description | Outer dense fiber protein. | |||||
|
ODFP_MOUSE
|
||||||
| NC score | 0.214751 (rank : 20) | θ value | 0.0148317 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61999 | Gene names | Odf1, Odfp | |||
|
Domain Architecture |

|
|||||
| Description | Outer dense fiber protein. | |||||
|
GAW2_HUMAN
|
||||||
| NC score | 0.024659 (rank : 21) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NRZ4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-W_3q26.32 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein (Gag P15); Capsid protein (Core shell protein) (Gag P30)]. | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.023428 (rank : 22) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
WBS14_MOUSE
|
||||||
| NC score | 0.018648 (rank : 23) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
CBPN_MOUSE
|
||||||
| NC score | 0.009885 (rank : 24) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJN5, Q91WM9 | Gene names | Cpn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase N catalytic chain precursor (EC 3.4.17.3) (CPN) (Carboxypeptidase N polypeptide 1) (Carboxypeptidase N small subunit). | |||||
|
MMP17_HUMAN
|
||||||
| NC score | 0.008829 (rank : 25) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULZ9, Q14850 | Gene names | MMP17, MT4MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
AKAP2_MOUSE
|
||||||
| NC score | 0.008421 (rank : 26) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.007367 (rank : 27) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||