Please be patient as the page loads
|
GSDM1_MOUSE
|
||||||
| SwissProt Accessions | Q9EST1 | Gene names | Gsdm1, Gsdm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gasdermin. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GSDM1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992123 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96QA5, Q8N1M6 | Gene names | GSDM1, GSDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gasdermin. | |||||
|
GSDM1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9EST1 | Gene names | Gsdm1, Gsdm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gasdermin. | |||||
|
MLZE_HUMAN
|
||||||
| θ value | 1.56636e-44 (rank : 3) | NC score | 0.856842 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BYG8, Q5XKF3, Q6P494 | Gene names | MLZE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-derived leucine zipper-containing extranuclear factor. | |||||
|
GSDC1_MOUSE
|
||||||
| θ value | 4.2656e-42 (rank : 4) | NC score | 0.848753 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D8T2, Q3TBD9 | Gene names | Gsdmdc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gasdermin domain-containing protein 1. | |||||
|
GSDC1_HUMAN
|
||||||
| θ value | 3.4976e-36 (rank : 5) | NC score | 0.843448 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P57764, Q96Q98 | Gene names | GSDMDC1, DFNA5L | |||
|
Domain Architecture |
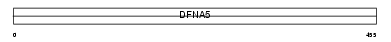
|
|||||
| Description | Gasdermin domain-containing protein 1. | |||||
|
PJVK_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 6) | NC score | 0.412691 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q0ZLH2 | Gene names | Pjvk, Dfnb59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pejvakin (Autosomal recessive deafness type 59 protein homolog). | |||||
|
PJVK_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 7) | NC score | 0.404382 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q0ZLH3 | Gene names | PJVK, DFNB59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pejvakin (Autosomal recessive deafness type 59 protein). | |||||
|
LAMB1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 8) | NC score | 0.022135 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
CEP70_MOUSE
|
||||||
| θ value | 0.813845 (rank : 9) | NC score | 0.028534 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6IQY5, Q9CRL9, Q9CTS4, Q9JIC1 | Gene names | Cep70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 70 kDa (Cep70 protein). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 10) | NC score | 0.015350 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
GOGA2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 11) | NC score | 0.017052 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 12) | NC score | 0.011006 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
LAMB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 13) | NC score | 0.018662 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
IPP4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.031519 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14990, Q5H958 | Gene names | PPP1R2P9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type-1 protein phosphatase inhibitor 4 (I-4) (Protein phosphatase 1, regulatory subunit 2 pseudogene 9). | |||||
|
AASS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.021118 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UDR5, O95462 | Gene names | AASS | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-aminoadipic semialdehyde synthase, mitochondrial precursor (LKR/SDH) [Includes: Lysine ketoglutarate reductase (EC 1.5.1.8) (LOR) (LKR); Saccharopine dehydrogenase (EC 1.5.1.9) (SDH)]. | |||||
|
APXL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.017940 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
TRI39_HUMAN
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.005634 (rank : 24) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCM9, Q76BL3, Q8IYT9, Q96IB6 | Gene names | TRIM39, RNF23, TFP | |||
|
Domain Architecture |
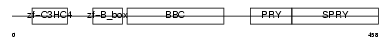
|
|||||
| Description | Tripartite motif-containing protein 39 (RING finger protein 23) (Testis-abundant finger protein). | |||||
|
VP13B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.021408 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z7G8, Q709C6, Q709C7, Q7Z7G4, Q7Z7G5, Q7Z7G6, Q7Z7G7, Q8NB77, Q9NWV1, Q9Y4E7 | Gene names | VPS13B, CHS1, COH1, KIAA0532 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 13B (Cohen syndrome protein 1). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.009531 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
SAS6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.012287 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UVJ0, Q8N3K0 | Gene names | SASS6, SAS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog (HsSAS-6). | |||||
|
CLGN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.008251 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P52194, Q9D2K5 | Gene names | Clgn, Meg1 | |||
|
Domain Architecture |
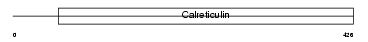
|
|||||
| Description | Calmegin precursor (MEG 1 antigen) (Calnexin-T) (A2/6). | |||||
|
TRAIP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.011715 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
DFNA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.159850 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60443, O14590, Q9UBV3 | Gene names | DFNA5, ICERE1 | |||
|
Domain Architecture |
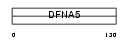
|
|||||
| Description | Non-syndromic hearing impairment protein 5 (Inversely correlated with estrogen receptor expression 1) (ICERE-1). | |||||
|
DFNA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 24) | NC score | 0.175826 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2D3 | Gene names | Dfna5, Dfna5h | |||
|
Domain Architecture |
|
|||||
| Description | Non-syndromic hearing impairment protein 5 homolog. | |||||
|
GSDM1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9EST1 | Gene names | Gsdm1, Gsdm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gasdermin. | |||||
|
GSDM1_HUMAN
|
||||||
| NC score | 0.992123 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96QA5, Q8N1M6 | Gene names | GSDM1, GSDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gasdermin. | |||||
|
MLZE_HUMAN
|
||||||
| NC score | 0.856842 (rank : 3) | θ value | 1.56636e-44 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BYG8, Q5XKF3, Q6P494 | Gene names | MLZE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-derived leucine zipper-containing extranuclear factor. | |||||
|
GSDC1_MOUSE
|
||||||
| NC score | 0.848753 (rank : 4) | θ value | 4.2656e-42 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D8T2, Q3TBD9 | Gene names | Gsdmdc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gasdermin domain-containing protein 1. | |||||
|
GSDC1_HUMAN
|
||||||
| NC score | 0.843448 (rank : 5) | θ value | 3.4976e-36 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P57764, Q96Q98 | Gene names | GSDMDC1, DFNA5L | |||
|
Domain Architecture |

|
|||||
| Description | Gasdermin domain-containing protein 1. | |||||
|
PJVK_MOUSE
|
||||||
| NC score | 0.412691 (rank : 6) | θ value | 5.62301e-10 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q0ZLH2 | Gene names | Pjvk, Dfnb59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pejvakin (Autosomal recessive deafness type 59 protein homolog). | |||||
|
PJVK_HUMAN
|
||||||
| NC score | 0.404382 (rank : 7) | θ value | 3.64472e-09 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q0ZLH3 | Gene names | PJVK, DFNB59 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pejvakin (Autosomal recessive deafness type 59 protein). | |||||
|
DFNA5_MOUSE
|
||||||
| NC score | 0.175826 (rank : 8) | θ value | θ > 10 (rank : 24) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2D3 | Gene names | Dfna5, Dfna5h | |||
|
Domain Architecture |

|
|||||
| Description | Non-syndromic hearing impairment protein 5 homolog. | |||||
|
DFNA5_HUMAN
|
||||||
| NC score | 0.159850 (rank : 9) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60443, O14590, Q9UBV3 | Gene names | DFNA5, ICERE1 | |||
|
Domain Architecture |

|
|||||
| Description | Non-syndromic hearing impairment protein 5 (Inversely correlated with estrogen receptor expression 1) (ICERE-1). | |||||
|
IPP4_HUMAN
|
||||||
| NC score | 0.031519 (rank : 10) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14990, Q5H958 | Gene names | PPP1R2P9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type-1 protein phosphatase inhibitor 4 (I-4) (Protein phosphatase 1, regulatory subunit 2 pseudogene 9). | |||||
|
CEP70_MOUSE
|
||||||
| NC score | 0.028534 (rank : 11) | θ value | 0.813845 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6IQY5, Q9CRL9, Q9CTS4, Q9JIC1 | Gene names | Cep70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 70 kDa (Cep70 protein). | |||||
|
LAMB1_HUMAN
|
||||||
| NC score | 0.022135 (rank : 12) | θ value | 0.163984 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
VP13B_HUMAN
|
||||||
| NC score | 0.021408 (rank : 13) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z7G8, Q709C6, Q709C7, Q7Z7G4, Q7Z7G5, Q7Z7G6, Q7Z7G7, Q8NB77, Q9NWV1, Q9Y4E7 | Gene names | VPS13B, CHS1, COH1, KIAA0532 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 13B (Cohen syndrome protein 1). | |||||
|
AASS_HUMAN
|
||||||
| NC score | 0.021118 (rank : 14) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UDR5, O95462 | Gene names | AASS | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-aminoadipic semialdehyde synthase, mitochondrial precursor (LKR/SDH) [Includes: Lysine ketoglutarate reductase (EC 1.5.1.8) (LOR) (LKR); Saccharopine dehydrogenase (EC 1.5.1.9) (SDH)]. | |||||
|
LAMB1_MOUSE
|
||||||
| NC score | 0.018662 (rank : 15) | θ value | 2.36792 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.017940 (rank : 16) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
GOGA2_MOUSE
|
||||||
| NC score | 0.017052 (rank : 17) | θ value | 1.38821 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.015350 (rank : 18) | θ value | 1.06291 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
SAS6_HUMAN
|
||||||
| NC score | 0.012287 (rank : 19) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6UVJ0, Q8N3K0 | Gene names | SASS6, SAS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog (HsSAS-6). | |||||
|
TRAIP_HUMAN
|
||||||
| NC score | 0.011715 (rank : 20) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.011006 (rank : 21) | θ value | 2.36792 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.009531 (rank : 22) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CLGN_MOUSE
|
||||||
| NC score | 0.008251 (rank : 23) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P52194, Q9D2K5 | Gene names | Clgn, Meg1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmegin precursor (MEG 1 antigen) (Calnexin-T) (A2/6). | |||||
|
TRI39_HUMAN
|
||||||
| NC score | 0.005634 (rank : 24) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCM9, Q76BL3, Q8IYT9, Q96IB6 | Gene names | TRIM39, RNF23, TFP | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 39 (RING finger protein 23) (Testis-abundant finger protein). | |||||