Please be patient as the page loads
|
ESPL1_HUMAN
|
||||||
| SwissProt Accessions | Q14674 | Gene names | ESPL1, ESP1, KIAA0165 | |||
|
Domain Architecture |

|
|||||
| Description | Separin (EC 3.4.22.49) (Separase) (Caspase-like protein ESPL1) (Extra spindle poles-like 1 protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ESPL1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q14674 | Gene names | ESPL1, ESP1, KIAA0165 | |||
|
Domain Architecture |

|
|||||
| Description | Separin (EC 3.4.22.49) (Separase) (Caspase-like protein ESPL1) (Extra spindle poles-like 1 protein). | |||||
|
ESPL1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.972047 (rank : 2) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60330 | Gene names | Espl1, Esp1, Kiaa0165 | |||
|
Domain Architecture |

|
|||||
| Description | Separin (EC 3.4.22.49) (Separase) (Caspase-like protein ESPL1) (Extra spindle poles-like 1 protein). | |||||
|
GASP1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 3) | NC score | 0.047049 (rank : 5) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5U4C1, Q8BKR8, Q8BUN4, Q8BYK9, Q8CHF4, Q8R095 | Gene names | Gprasp1, Kiaa0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 4) | NC score | 0.055450 (rank : 4) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
FKBP4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 5) | NC score | 0.036604 (rank : 8) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30416, Q3TVC9 | Gene names | Fkbp4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.365318 (rank : 6) | NC score | 0.028524 (rank : 12) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
CO1A1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 7) | NC score | 0.026178 (rank : 16) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
PNOC_MOUSE
|
||||||
| θ value | 0.47712 (rank : 8) | NC score | 0.060843 (rank : 3) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64387, Q61105, Q61938 | Gene names | Pnoc, Npnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Nociceptin precursor (N23K/N27K) [Contains: Neuropeptide 1; Nociceptin (Orphanin FQ) (PPNOC); Neuropeptide 2]. | |||||
|
FKBP4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 9) | NC score | 0.033725 (rank : 9) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02790, Q9UCP1, Q9UCV7 | Gene names | FKBP4 | |||
|
Domain Architecture |
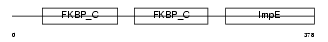
|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
KIF1A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 10) | NC score | 0.018212 (rank : 46) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12756 | Gene names | KIF1A, ATSV | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1A (Axonal transporter of synaptic vesicles). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 11) | NC score | 0.023333 (rank : 28) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
CO1A1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 12) | NC score | 0.025604 (rank : 18) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11087, Q53WT0, Q60635, Q61367, Q61427, Q63919, Q6PCL3, Q810J9 | Gene names | Col1a1, Cola1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
CYLN2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 13) | NC score | 0.014310 (rank : 58) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
DDX51_HUMAN
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.017790 (rank : 50) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
GP179_HUMAN
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.033197 (rank : 10) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.023789 (rank : 26) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
BSPRY_MOUSE
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.021432 (rank : 32) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80YW5, Q3TU74, Q8BZF0, Q99KV7, Q9ER70 | Gene names | Bspry | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B box and SPRY domain-containing protein. | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.031364 (rank : 11) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
KIF6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.017232 (rank : 51) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZMV9, Q2MDE3, Q2MDE4, Q5T8J6, Q6ZWE3, Q86T87, Q8WTV4 | Gene names | KIF6, C6orf102 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF6. | |||||
|
O10Q1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.001612 (rank : 95) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NGQ4 | Gene names | OR10Q1 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 10Q1. | |||||
|
ZFP38_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.006281 (rank : 87) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 766 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y5A6, Q9H0B5 | Gene names | ZNF38, ZFP38, ZSCAN21 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 38 homolog (Zfp-38) (Zinc finger and SCAN domain- containing protein 21) (NY-REN-21 antigen). | |||||
|
FOXO4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.020786 (rank : 34) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P98177, O43821, Q13720, Q3KPF1, Q8TDK9 | Gene names | MLLT7, AFX, AFX1, FOXO4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Forkhead box protein O4). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.023687 (rank : 27) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
NADAP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.041270 (rank : 7) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BWU0, Q4KMT1, Q4KMX0, Q7Z5Q9, Q9NVN2 | Gene names | SLC4A1AP, HLC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kanadaptin (Kidney anion exchanger adapter protein) (Solute carrier family 4 anion exchanger member 1 adapter protein) (Lung cancer oncogene 3 protein). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.024579 (rank : 24) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.006258 (rank : 88) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ZN263_HUMAN
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.005321 (rank : 90) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14978, O43387, Q96H95 | Gene names | ZNF263, FPM315 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 263 (Zinc finger protein FPM315). | |||||
|
FOXB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.016146 (rank : 55) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99853, O60652, O75917 | Gene names | FOXB1, FKH5 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein B1 (Transcription factor FKH-5). | |||||
|
FOXB1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.016473 (rank : 53) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64732, O08753 | Gene names | Foxb1, Fkh5, Foxb1a, Foxb1b, Mf3 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein B1 (Transcription factor FKH-5). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.019002 (rank : 38) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
DLGP4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.021291 (rank : 33) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2H0, Q5T2Y4, Q5T2Y5, Q9H137, Q9H138, Q9H1L7 | Gene names | DLGAP4, DAP4, KIAA0964 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large-associated protein 4 (DAP-4) (SAP90/PSD-95-associated protein 4) (SAPAP4) (PSD-95/SAP90-binding protein 4). | |||||
|
GLI2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.006960 (rank : 85) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.022924 (rank : 29) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.025137 (rank : 19) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
ZN397_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.004910 (rank : 93) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NF99, Q9BRM2 | Gene names | ZNF397, ZSCAN15 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 397 (Zinc finger and SCAN domain-containing protein 15). | |||||
|
ATX7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.024993 (rank : 22) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.044886 (rank : 6) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.021571 (rank : 31) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.018439 (rank : 44) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.024914 (rank : 23) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TRI66_MOUSE
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.023865 (rank : 25) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
U383_MOUSE
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.025683 (rank : 17) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D2Q2, Q3V1K3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative methyltransferase UPF0383 (EC 2.1.1.-). | |||||
|
ZN165_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.005460 (rank : 89) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 734 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49910 | Gene names | ZNF165, ZPF165, ZSCAN7 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 165 (LD65) (Zinc finger and SCAN domain-containing protein 7). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.016523 (rank : 52) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.017809 (rank : 49) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.019494 (rank : 37) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PUR4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.028284 (rank : 13) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15067 | Gene names | PFAS, KIAA0361 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoribosylformylglycinamidine synthase (EC 6.3.5.3) (FGAM synthase) (FGAMS) (Formylglycinamide ribotide amidotransferase) (FGARAT) (Formylglycinamide ribotide synthetase). | |||||
|
SYN2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.018545 (rank : 42) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q64332, Q6NZR0, Q9QWV7 | Gene names | Syn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
TRAF2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.025036 (rank : 21) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P39429, O54896 | Gene names | Traf2 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 2. | |||||
|
WRN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.027700 (rank : 15) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.017990 (rank : 47) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
AP3D1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.017857 (rank : 48) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
CBX7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.016203 (rank : 54) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CO3A1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.019871 (rank : 36) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
CO4A5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.015310 (rank : 56) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
COBA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.015028 (rank : 57) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
CTDP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.025071 (rank : 20) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
PACS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.018629 (rank : 41) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K212, Q6P5H8 | Gene names | Pacs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphofurin acidic cluster sorting protein 1 (PACS-1). | |||||
|
PPE2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.011087 (rank : 69) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35385 | Gene names | Ppef2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 2 (EC 3.1.3.16) (PPEF-2). | |||||
|
SELV_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.028135 (rank : 14) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
TTLL5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.012617 (rank : 65) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
ZN446_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.007644 (rank : 79) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NWS9 | Gene names | ZNF446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 446. | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.013406 (rank : 60) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
APXL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.020778 (rank : 35) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
BRWD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.007122 (rank : 83) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.009436 (rank : 75) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.018489 (rank : 43) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
KIF1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.013913 (rank : 59) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P33173, Q61770 | Gene names | Kif1a, Atsv, Kif1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1A (Axonal transporter of synaptic vesicles). | |||||
|
KLHL6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.004046 (rank : 94) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6V595 | Gene names | Klhl6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 6. | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.022632 (rank : 30) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
RUFY1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.008547 (rank : 78) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
S12A5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.007099 (rank : 84) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91V14, Q9Z0M7 | Gene names | Slc12a5, Kcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (mKCC2). | |||||
|
STIM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.010623 (rank : 71) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.013266 (rank : 61) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
TAP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.005209 (rank : 91) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03519, Q9UQ83 | Gene names | TAP2, ABCB3, PSF2, RING11, Y1 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen peptide transporter 2 (APT2) (ATP-binding cassette sub-family B member 3) (Peptide transporter TAP2) (Peptide transporter PSF2) (Peptide supply factor 2) (PSF-2) (Peptide transporter involved in antigen processing 2). | |||||
|
TULP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.018642 (rank : 40) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
TULP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.018761 (rank : 39) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIL5, Q8CA75, Q922C2 | Gene names | Tulp4, Tusp | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
WDHD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.009164 (rank : 76) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59328, Q6P408 | Gene names | Wdhd1, And1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
ZN193_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.004974 (rank : 92) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15535 | Gene names | ZNF193, ZSCAN9 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 193 (PRD51) (Zinc finger and SCAN domain- containing protein 9). | |||||
|
ZN687_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.007193 (rank : 82) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
ARHG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.009775 (rank : 74) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
BAT3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.018271 (rank : 45) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
BUB1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.011030 (rank : 70) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60566, O60501, O60627, O60758, O75389, Q96KM4 | Gene names | BUB1B, BUBR1, MAD3L | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (hBUBR1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L) (SSK1). | |||||
|
CABP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.007211 (rank : 81) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
COEA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.008956 (rank : 77) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80X19, Q8C6X3, Q9WV05 | Gene names | Col14a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor. | |||||
|
DLEC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.013154 (rank : 63) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y238, Q9NSW0, Q9NTG5 | Gene names | DLEC1, DLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in lung and esophageal cancer protein 1 (Deleted in lung cancer protein 1) (DLC-1). | |||||
|
HIP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.006536 (rank : 86) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
JIP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.012780 (rank : 64) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
LBN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.010244 (rank : 72) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UK5, Q86YT3, Q86YT4, Q8NG49 | Gene names | EVC2, LBN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Limbin. | |||||
|
LYST_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.011160 (rank : 68) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
NIBL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.013254 (rank : 62) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96TA1, Q9BUS2, Q9NT35 | Gene names | C9orf88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban-like protein (Meg-3). | |||||
|
PKHG4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.012289 (rank : 67) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q58EX7, Q4G0J8, Q4H485, Q56A69, Q9H7K4, Q9UFW0 | Gene names | PLEKHG4, PRTPHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Puratrophin-1 (Pleckstrin homology domain-containing family G member 4) (Purkinje cell atrophy-associated protein 1). | |||||
|
SPTN5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.007430 (rank : 80) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
TB22A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.009870 (rank : 73) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R5A6, Q3U268, Q3U3T0, Q8CA49 | Gene names | Tbc1d22a, D15Ertd781e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 22A. | |||||
|
ZCH11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.012482 (rank : 66) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
ESPL1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q14674 | Gene names | ESPL1, ESP1, KIAA0165 | |||
|
Domain Architecture |

|
|||||
| Description | Separin (EC 3.4.22.49) (Separase) (Caspase-like protein ESPL1) (Extra spindle poles-like 1 protein). | |||||
|
ESPL1_MOUSE
|
||||||
| NC score | 0.972047 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60330 | Gene names | Espl1, Esp1, Kiaa0165 | |||
|
Domain Architecture |

|
|||||
| Description | Separin (EC 3.4.22.49) (Separase) (Caspase-like protein ESPL1) (Extra spindle poles-like 1 protein). | |||||
|
PNOC_MOUSE
|
||||||
| NC score | 0.060843 (rank : 3) | θ value | 0.47712 (rank : 8) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64387, Q61105, Q61938 | Gene names | Pnoc, Npnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Nociceptin precursor (N23K/N27K) [Contains: Neuropeptide 1; Nociceptin (Orphanin FQ) (PPNOC); Neuropeptide 2]. | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.055450 (rank : 4) | θ value | 0.125558 (rank : 4) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
GASP1_MOUSE
|
||||||
| NC score | 0.047049 (rank : 5) | θ value | 0.0330416 (rank : 3) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5U4C1, Q8BKR8, Q8BUN4, Q8BYK9, Q8CHF4, Q8R095 | Gene names | Gprasp1, Kiaa0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.044886 (rank : 6) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
NADAP_HUMAN
|
||||||
| NC score | 0.041270 (rank : 7) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BWU0, Q4KMT1, Q4KMX0, Q7Z5Q9, Q9NVN2 | Gene names | SLC4A1AP, HLC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kanadaptin (Kidney anion exchanger adapter protein) (Solute carrier family 4 anion exchanger member 1 adapter protein) (Lung cancer oncogene 3 protein). | |||||
|
FKBP4_MOUSE
|
||||||
| NC score | 0.036604 (rank : 8) | θ value | 0.279714 (rank : 5) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30416, Q3TVC9 | Gene names | Fkbp4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP4_HUMAN
|
||||||
| NC score | 0.033725 (rank : 9) | θ value | 0.62314 (rank : 9) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02790, Q9UCP1, Q9UCV7 | Gene names | FKBP4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.033197 (rank : 10) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.031364 (rank : 11) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.028524 (rank : 12) | θ value | 0.365318 (rank : 6) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PUR4_HUMAN
|
||||||
| NC score | 0.028284 (rank : 13) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15067 | Gene names | PFAS, KIAA0361 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoribosylformylglycinamidine synthase (EC 6.3.5.3) (FGAM synthase) (FGAMS) (Formylglycinamide ribotide amidotransferase) (FGARAT) (Formylglycinamide ribotide synthetase). | |||||
|
SELV_HUMAN
|
||||||
| NC score | 0.028135 (rank : 14) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
WRN_MOUSE
|
||||||
| NC score | 0.027700 (rank : 15) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
CO1A1_HUMAN
|
||||||
| NC score | 0.026178 (rank : 16) | θ value | 0.47712 (rank : 7) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
U383_MOUSE
|
||||||
| NC score | 0.025683 (rank : 17) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D2Q2, Q3V1K3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative methyltransferase UPF0383 (EC 2.1.1.-). | |||||
|
CO1A1_MOUSE
|
||||||
| NC score | 0.025604 (rank : 18) | θ value | 0.813845 (rank : 12) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11087, Q53WT0, Q60635, Q61367, Q61427, Q63919, Q6PCL3, Q810J9 | Gene names | Col1a1, Cola1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.025137 (rank : 19) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
CTDP1_HUMAN
|
||||||
| NC score | 0.025071 (rank : 20) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
TRAF2_MOUSE
|
||||||
| NC score | 0.025036 (rank : 21) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P39429, O54896 | Gene names | Traf2 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 2. | |||||
|
ATX7_HUMAN
|
||||||
| NC score | 0.024993 (rank : 22) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.024914 (rank : 23) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.024579 (rank : 24) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.023865 (rank : 25) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.023789 (rank : 26) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.023687 (rank : 27) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.023333 (rank : 28) | θ value | 0.62314 (rank : 11) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.022924 (rank : 29) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.022632 (rank : 30) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.021571 (rank : 31) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
BSPRY_MOUSE
|
||||||
| NC score | 0.021432 (rank : 32) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80YW5, Q3TU74, Q8BZF0, Q99KV7, Q9ER70 | Gene names | Bspry | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B box and SPRY domain-containing protein. | |||||
|
DLGP4_HUMAN
|
||||||
| NC score | 0.021291 (rank : 33) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2H0, Q5T2Y4, Q5T2Y5, Q9H137, Q9H138, Q9H1L7 | Gene names | DLGAP4, DAP4, KIAA0964 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large-associated protein 4 (DAP-4) (SAP90/PSD-95-associated protein 4) (SAPAP4) (PSD-95/SAP90-binding protein 4). | |||||
|
FOXO4_HUMAN
|
||||||
| NC score | 0.020786 (rank : 34) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P98177, O43821, Q13720, Q3KPF1, Q8TDK9 | Gene names | MLLT7, AFX, AFX1, FOXO4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Forkhead box protein O4). | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.020778 (rank : 35) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
CO3A1_HUMAN
|
||||||
| NC score | 0.019871 (rank : 36) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.019494 (rank : 37) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.019002 (rank : 38) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
TULP4_MOUSE
|
||||||
| NC score | 0.018761 (rank : 39) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIL5, Q8CA75, Q922C2 | Gene names | Tulp4, Tusp | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
TULP4_HUMAN
|
||||||
| NC score | 0.018642 (rank : 40) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
PACS1_MOUSE
|
||||||
| NC score | 0.018629 (rank : 41) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K212, Q6P5H8 | Gene names | Pacs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphofurin acidic cluster sorting protein 1 (PACS-1). | |||||
|
SYN2_MOUSE
|
||||||
| NC score | 0.018545 (rank : 42) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q64332, Q6NZR0, Q9QWV7 | Gene names | Syn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.018489 (rank : 43) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.018439 (rank : 44) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.018271 (rank : 45) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
KIF1A_HUMAN
|
||||||
| NC score | 0.018212 (rank : 46) | θ value | 0.62314 (rank : 10) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12756 | Gene names | KIF1A, ATSV | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1A (Axonal transporter of synaptic vesicles). | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.017990 (rank : 47) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
AP3D1_MOUSE
|
||||||
| NC score | 0.017857 (rank : 48) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54774 | Gene names | Ap3d1, Ap3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin) (mBLVR1). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.017809 (rank : 49) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
DDX51_HUMAN
|
||||||
| NC score | 0.017790 (rank : 50) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
KIF6_HUMAN
|
||||||
| NC score | 0.017232 (rank : 51) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZMV9, Q2MDE3, Q2MDE4, Q5T8J6, Q6ZWE3, Q86T87, Q8WTV4 | Gene names | KIF6, C6orf102 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF6. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.016523 (rank : 52) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
FOXB1_MOUSE
|
||||||
| NC score | 0.016473 (rank : 53) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64732, O08753 | Gene names | Foxb1, Fkh5, Foxb1a, Foxb1b, Mf3 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein B1 (Transcription factor FKH-5). | |||||
|
CBX7_HUMAN
|
||||||
| NC score | 0.016203 (rank : 54) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
FOXB1_HUMAN
|
||||||
| NC score | 0.016146 (rank : 55) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99853, O60652, O75917 | Gene names | FOXB1, FKH5 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein B1 (Transcription factor FKH-5). | |||||
|
CO4A5_HUMAN
|
||||||
| NC score | 0.015310 (rank : 56) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
COBA2_MOUSE
|
||||||
| NC score | 0.015028 (rank : 57) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
CYLN2_HUMAN
|
||||||
| NC score | 0.014310 (rank : 58) | θ value | 0.813845 (rank : 13) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
KIF1A_MOUSE
|
||||||
| NC score | 0.013913 (rank : 59) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P33173, Q61770 | Gene names | Kif1a, Atsv, Kif1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1A (Axonal transporter of synaptic vesicles). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.013406 (rank : 60) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.013266 (rank : 61) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
NIBL_HUMAN
|
||||||
| NC score | 0.013254 (rank : 62) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96TA1, Q9BUS2, Q9NT35 | Gene names | C9orf88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban-like protein (Meg-3). | |||||
|
DLEC1_HUMAN
|
||||||
| NC score | 0.013154 (rank : 63) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y238, Q9NSW0, Q9NTG5 | Gene names | DLEC1, DLC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in lung and esophageal cancer protein 1 (Deleted in lung cancer protein 1) (DLC-1). | |||||
|
JIP1_HUMAN
|
||||||
| NC score | 0.012780 (rank : 64) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
TTLL5_HUMAN
|
||||||
| NC score | 0.012617 (rank : 65) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
ZCH11_HUMAN
|
||||||
| NC score | 0.012482 (rank : 66) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
PKHG4_HUMAN
|
||||||
| NC score | 0.012289 (rank : 67) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q58EX7, Q4G0J8, Q4H485, Q56A69, Q9H7K4, Q9UFW0 | Gene names | PLEKHG4, PRTPHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Puratrophin-1 (Pleckstrin homology domain-containing family G member 4) (Purkinje cell atrophy-associated protein 1). | |||||
|
LYST_MOUSE
|
||||||
| NC score | 0.011160 (rank : 68) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
PPE2_MOUSE
|
||||||
| NC score | 0.011087 (rank : 69) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35385 | Gene names | Ppef2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 2 (EC 3.1.3.16) (PPEF-2). | |||||
|
BUB1B_HUMAN
|
||||||
| NC score | 0.011030 (rank : 70) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60566, O60501, O60627, O60758, O75389, Q96KM4 | Gene names | BUB1B, BUBR1, MAD3L | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (hBUBR1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L) (SSK1). | |||||
|
STIM1_HUMAN
|
||||||
| NC score | 0.010623 (rank : 71) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
LBN_HUMAN
|
||||||
| NC score | 0.010244 (rank : 72) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UK5, Q86YT3, Q86YT4, Q8NG49 | Gene names | EVC2, LBN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Limbin. | |||||
|
TB22A_MOUSE
|
||||||
| NC score | 0.009870 (rank : 73) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R5A6, Q3U268, Q3U3T0, Q8CA49 | Gene names | Tbc1d22a, D15Ertd781e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 22A. | |||||
|
ARHG1_MOUSE
|
||||||
| NC score | 0.009775 (rank : 74) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.009436 (rank : 75) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
WDHD1_MOUSE
|
||||||
| NC score | 0.009164 (rank : 76) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59328, Q6P408 | Gene names | Wdhd1, And1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
COEA1_MOUSE
|
||||||
| NC score | 0.008956 (rank : 77) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80X19, Q8C6X3, Q9WV05 | Gene names | Col14a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor. | |||||
|
RUFY1_MOUSE
|
||||||
| NC score | 0.008547 (rank : 78) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
ZN446_HUMAN
|
||||||
| NC score | 0.007644 (rank : 79) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NWS9 | Gene names | ZNF446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 446. | |||||
|
SPTN5_HUMAN
|
||||||
| NC score | 0.007430 (rank : 80) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
CABP4_HUMAN
|
||||||
| NC score | 0.007211 (rank : 81) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
ZN687_MOUSE
|
||||||
| NC score | 0.007193 (rank : 82) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
BRWD1_MOUSE
|
||||||
| NC score | 0.007122 (rank : 83) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
S12A5_MOUSE
|
||||||
| NC score | 0.007099 (rank : 84) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91V14, Q9Z0M7 | Gene names | Slc12a5, Kcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (mKCC2). | |||||
|
GLI2_HUMAN
|
||||||
| NC score | 0.006960 (rank : 85) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.006536 (rank : 86) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
ZFP38_HUMAN
|
||||||
| NC score | 0.006281 (rank : 87) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 766 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y5A6, Q9H0B5 | Gene names | ZNF38, ZFP38, ZSCAN21 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 38 homolog (Zfp-38) (Zinc finger and SCAN domain- containing protein 21) (NY-REN-21 antigen). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.006258 (rank : 88) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ZN165_HUMAN
|
||||||
| NC score | 0.005460 (rank : 89) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 734 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49910 | Gene names | ZNF165, ZPF165, ZSCAN7 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 165 (LD65) (Zinc finger and SCAN domain-containing protein 7). | |||||
|
ZN263_HUMAN
|
||||||
| NC score | 0.005321 (rank : 90) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14978, O43387, Q96H95 | Gene names | ZNF263, FPM315 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 263 (Zinc finger protein FPM315). | |||||
|
TAP2_HUMAN
|
||||||
| NC score | 0.005209 (rank : 91) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03519, Q9UQ83 | Gene names | TAP2, ABCB3, PSF2, RING11, Y1 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen peptide transporter 2 (APT2) (ATP-binding cassette sub-family B member 3) (Peptide transporter TAP2) (Peptide transporter PSF2) (Peptide supply factor 2) (PSF-2) (Peptide transporter involved in antigen processing 2). | |||||
|
ZN193_HUMAN
|
||||||
| NC score | 0.004974 (rank : 92) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15535 | Gene names | ZNF193, ZSCAN9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 193 (PRD51) (Zinc finger and SCAN domain- containing protein 9). | |||||
|
ZN397_HUMAN
|
||||||
| NC score | 0.004910 (rank : 93) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NF99, Q9BRM2 | Gene names | ZNF397, ZSCAN15 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 397 (Zinc finger and SCAN domain-containing protein 15). | |||||
|
KLHL6_MOUSE
|
||||||
| NC score | 0.004046 (rank : 94) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6V595 | Gene names | Klhl6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 6. | |||||
|
O10Q1_HUMAN
|
||||||
| NC score | 0.001612 (rank : 95) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NGQ4 | Gene names | OR10Q1 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 10Q1. | |||||