Please be patient as the page loads
|
DLGP4_HUMAN
|
||||||
| SwissProt Accessions | Q9Y2H0, Q5T2Y4, Q5T2Y5, Q9H137, Q9H138, Q9H1L7 | Gene names | DLGAP4, DAP4, KIAA0964 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large-associated protein 4 (DAP-4) (SAP90/PSD-95-associated protein 4) (SAPAP4) (PSD-95/SAP90-binding protein 4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DLGP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.961370 (rank : 3) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14490, O14489, P78335 | Gene names | DLGAP1, DAP1, GKAP | |||
|
Domain Architecture |

|
|||||
| Description | Disks large-associated protein 1 (DAP-1) (Guanylate kinase-associated protein) (hGKAP) (SAP90/PSD-95-associated protein 1) (SAPAP1) (PSD- 95/SAP90-binding protein 1). | |||||
|
DLGP4_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9Y2H0, Q5T2Y4, Q5T2Y5, Q9H137, Q9H138, Q9H1L7 | Gene names | DLGAP4, DAP4, KIAA0964 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large-associated protein 4 (DAP-4) (SAP90/PSD-95-associated protein 4) (SAPAP4) (PSD-95/SAP90-binding protein 4). | |||||
|
DLGP1_MOUSE
|
||||||
| θ value | 2.40406e-178 (rank : 3) | NC score | 0.961550 (rank : 2) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D415, Q52KF6, Q5DTK5, Q6P6N4, Q6XBF4, Q8BZL7, Q8BZQ1, Q8C0G0 | Gene names | Dlgap1, Kiaa4162 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 1 (DAP-1) (Guanylate kinase-associated protein) (SAP90/PSD-95-associated protein 1) (SAPAP1) (PSD-95/SAP90- binding protein 1). | |||||
|
DLGP2_MOUSE
|
||||||
| θ value | 1.74296e-136 (rank : 4) | NC score | 0.948310 (rank : 4) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BJ42, Q6XBF3 | Gene names | Dlgap2, Dap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 2 (DAP-2) (SAP90/PSD-95-associated protein 2) (SAPAP2) (PSD-95/SAP90-binding protein 2). | |||||
|
DLGP3_MOUSE
|
||||||
| θ value | 2.25659e-51 (rank : 5) | NC score | 0.883220 (rank : 5) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PFD5, Q6PDX0, Q6XBF2 | Gene names | Dlgap3, Dap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
DLGP3_HUMAN
|
||||||
| θ value | 3.84914e-51 (rank : 6) | NC score | 0.882698 (rank : 6) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95886, Q5TDD5, Q9H3X7 | Gene names | DLGAP3, DAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
DLG7_MOUSE
|
||||||
| θ value | 1.13037e-18 (rank : 7) | NC score | 0.610592 (rank : 7) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K4R9, Q6ZQL3, Q80VS7, Q8BUC8, Q8BZ79 | Gene names | Dlg7, Kiaa0008 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discs large homolog 7 (Hepatoma up-regulated protein homolog) (HURP). | |||||
|
DLG7_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 8) | NC score | 0.589245 (rank : 8) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15398, Q8NG58 | Gene names | DLG7, KIAA0008 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 7 (Hepatoma up-regulated protein) (HURP). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 9) | NC score | 0.027955 (rank : 23) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
ZN509_MOUSE
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.004879 (rank : 81) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
IKBE_MOUSE
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.016859 (rank : 44) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54910, Q3U686, Q9CZZ9, Q9D7U3 | Gene names | Nfkbie, Ikbe | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKB-E). | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.022961 (rank : 30) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.040746 (rank : 12) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
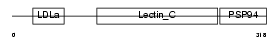
TARA_HUMAN
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.029966 (rank : 18) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
DPOLZ_MOUSE
|
||||||
| θ value | 0.279714 (rank : 15) | NC score | 0.031610 (rank : 17) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61493, Q9JMD6, Q9QWX6 | Gene names | Rev3l, Polz, Sez4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (Seizure-related protein 4). | |||||
|
ITA6_HUMAN
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.017088 (rank : 43) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23229, Q08443, Q14646, Q16508, Q9UN03 | Gene names | ITGA6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-6 precursor (VLA-6) (CD49f antigen) [Contains: Integrin alpha-6 heavy chain; Integrin alpha-6 light chain]. | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.034355 (rank : 16) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
MK07_MOUSE
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.003622 (rank : 86) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
FBX41_MOUSE
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.038159 (rank : 13) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6NS60, Q6P7W4, Q6ZPG1 | Gene names | Fbxo41, D6Ertd538e, Kiaa1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.013939 (rank : 53) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
PHLB2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.016629 (rank : 45) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
PTN13_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.010154 (rank : 66) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.024935 (rank : 27) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.037911 (rank : 14) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
MUCEN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.059695 (rank : 10) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R0H2, Q78KL2, Q9DCN9, Q9ULC1, Q9Z2I1 | Gene names | Emcn, Muc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endomucin precursor (Endomucin-1/2). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.058681 (rank : 11) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ZN174_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.004381 (rank : 84) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15697, Q9BQ34 | Gene names | ZNF174, ZSCAN8 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 174 (AW-1) (Zinc finger and SCAN domain-containing protein 8). | |||||
|
DNL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.028905 (rank : 20) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
EVI1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.003960 (rank : 85) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
FBX41_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.035354 (rank : 15) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
NID2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.008878 (rank : 72) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
PDLI5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.010716 (rank : 62) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CI51, Q9QYN0, Q9QYN1, Q9QYN2 | Gene names | Pdlim5, Enh | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
TOPRS_MOUSE
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.024595 (rank : 28) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80Z37, Q3U5B5, Q3UPZ1, Q3USN3, Q3UZ55, Q8BXP2, Q8CFF5, Q8CGC8, Q920L3 | Gene names | Topors | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
HAP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.019167 (rank : 38) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35668, O35636 | Gene names | Hap1 | |||
|
Domain Architecture |
|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1). | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.017652 (rank : 41) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
INAR2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.029225 (rank : 19) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35664, O35238, O35663, O35983 | Gene names | Ifnar2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-alpha/beta receptor beta chain precursor (IFN-alpha-REC) (Type I interferon receptor) (IFN-R) (Interferon alpha/beta receptor 2). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.015740 (rank : 48) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SAMN1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.025510 (rank : 25) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57725, Q8C6S2 | Gene names | Samsn1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM domain-containing protein SAMSN-1 (SAM domain, SH3 domain and nuclear localization signals protein 1). | |||||
|
SASH1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.023406 (rank : 29) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O94885, Q8TAI0, Q9H7R7 | Gene names | SASH1, KIAA0790, PEPE1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1 (Proline-glutamate repeat- containing protein). | |||||
|
SREC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.009510 (rank : 68) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
ESPL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.021291 (rank : 34) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14674 | Gene names | ESPL1, ESP1, KIAA0165 | |||
|
Domain Architecture |

|
|||||
| Description | Separin (EC 3.4.22.49) (Separase) (Caspase-like protein ESPL1) (Extra spindle poles-like 1 protein). | |||||
|
PLIN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.021868 (rank : 32) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60240, Q8N5Y6 | Gene names | PLIN | |||
|
Domain Architecture |

|
|||||
| Description | Perilipin (PERI) (Lipid droplet-associated protein). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.019968 (rank : 36) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
BRWD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.010676 (rank : 64) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
CARD6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.028145 (rank : 22) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BX69 | Gene names | CARD6 | |||
|
Domain Architecture |
|
|||||
| Description | Caspase recruitment domain-containing protein 6. | |||||
|
CTGE5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.012188 (rank : 57) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.028401 (rank : 21) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
RPGF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.015710 (rank : 49) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.064559 (rank : 9) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ZFP95_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.001575 (rank : 90) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2L8 | Gene names | ZFP95, KIAA1015 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 95 homolog (Zfp-95). | |||||
|
CTGE6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.012011 (rank : 59) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86UF2 | Gene names | CTAGE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-6 (cTAGE family member 6). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.019570 (rank : 37) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.026366 (rank : 24) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
SIPA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.012965 (rank : 56) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96FS4, O14518, O60484, O60618 | Gene names | SIPA1, SPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1) (p130 SPA-1). | |||||
|
SON_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.013803 (rank : 54) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TACC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.011790 (rank : 60) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
AKAP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.016072 (rank : 46) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.022112 (rank : 31) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.021484 (rank : 33) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
SAPS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.018505 (rank : 39) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.025419 (rank : 26) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
VPS53_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.014884 (rank : 51) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CCB4, Q8C7U0, Q8CIA1, Q9CX82, Q9D6T7 | Gene names | Vps53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 53. | |||||
|
ACHA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.003561 (rank : 88) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15822, Q9HAQ3 | Gene names | CHRNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal acetylcholine receptor protein subunit alpha-2 precursor. | |||||
|
ARHG1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.007329 (rank : 77) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92888, O00513, Q8N4J4, Q96BF4, Q96F17, Q9BSB1 | Gene names | ARHGEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (p115-RhoGEF) (p115RhoGEF) (115 kDa guanine nucleotide exchange factor) (Sub1.5). | |||||
|
CO002_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.015263 (rank : 50) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
E2F3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.011771 (rank : 61) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00716, Q15000, Q9BZ44 | Gene names | E2F3, KIAA0075 | |||
|
Domain Architecture |
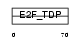
|
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
IDD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.010703 (rank : 63) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98154, Q61844 | Gene names | Dgcr2, Dgsc, Idd, Sez-12, Sez12 | |||
|
Domain Architecture |
|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor (Seizure-related membrane-bound adhesion protein). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.017215 (rank : 42) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.003584 (rank : 87) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
PHF12_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.009881 (rank : 67) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PJA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.010327 (rank : 65) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NG27, Q8NG28, Q9HAC1 | Gene names | PJA1, RNF70 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin protein ligase Praja1 (EC 6.3.2.-) (RING finger protein 70). | |||||
|
REXO4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.013297 (rank : 55) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PAQ4, Q3UU17, Q8C524 | Gene names | Rexo4, Gm111, Pmc2, Xpmc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 4 (EC 3.1.-.-) (Exonuclease XPMC2) (Prevents mitotic catastrophe 2 protein homolog). | |||||
|
TROAP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.020191 (rank : 35) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
WDR20_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.009058 (rank : 71) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TBZ3, Q8WXX2, Q9UF86 | Gene names | WDR20 | |||
|
Domain Architecture |
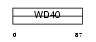
|
|||||
| Description | WD repeat protein 20 (DMR protein). | |||||
|
CCD39_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.004651 (rank : 83) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CK024_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.017817 (rank : 40) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
FBLN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.003551 (rank : 89) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
GASP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.008493 (rank : 74) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96D09, Q8NAB4 | Gene names | GPRASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.014167 (rank : 52) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
NAGS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.012087 (rank : 58) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R4H7, Q8C6G6, Q8CI77, Q8K1R8 | Gene names | Nags | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglutamate synthase, mitochondrial precursor (EC 2.3.1.1) (Amino-acid acetyltransferase) [Contains: N-acetylglutamate synthase long form; N-acetylglutamate synthase short form; N-acetylglutamate synthase conserved domain form]. | |||||
|
OXR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.009506 (rank : 69) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N573, Q3LIB5, Q6ZVK9, Q8N8V0, Q9H266, Q9NWC7 | Gene names | OXR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1. | |||||
|
QSCN6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.008530 (rank : 73) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BND5, Q3TDY9, Q3TE19, Q3TR29, Q3UEL4, Q8K041, Q9DBL6 | Gene names | Qscn6, Sox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfhydryl oxidase 1 precursor (EC 1.8.3.2) (Quiescin Q6) (Skin sulfhydryl oxidase) (mSOx). | |||||
|
REPS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.009465 (rank : 70) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96D71, Q8NDR7, Q8WU62, Q9BXY9 | Gene names | REPS1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.004878 (rank : 82) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
RTP3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.015880 (rank : 47) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.007167 (rank : 79) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
SYNJ2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.008022 (rank : 75) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15056, Q5TA13, Q5TA16, Q5TA19, Q86XK0, Q8IZA8, Q9H226 | Gene names | SYNJ2, KIAA0348 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
TF7L2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.005621 (rank : 80) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
TIAM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.007303 (rank : 78) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
ZF106_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.007362 (rank : 76) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H2Y7, Q6NSD9, Q6PEK1, Q86T43, Q86T45, Q86T50, Q86T58, Q86TA9, Q96M37, Q9H7B8 | Gene names | ZFP106, ZNF474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 homolog (Zfp-106) (Zinc finger protein 474). | |||||
|
DLGP4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9Y2H0, Q5T2Y4, Q5T2Y5, Q9H137, Q9H138, Q9H1L7 | Gene names | DLGAP4, DAP4, KIAA0964 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large-associated protein 4 (DAP-4) (SAP90/PSD-95-associated protein 4) (SAPAP4) (PSD-95/SAP90-binding protein 4). | |||||
|
DLGP1_MOUSE
|
||||||
| NC score | 0.961550 (rank : 2) | θ value | 2.40406e-178 (rank : 3) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D415, Q52KF6, Q5DTK5, Q6P6N4, Q6XBF4, Q8BZL7, Q8BZQ1, Q8C0G0 | Gene names | Dlgap1, Kiaa4162 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 1 (DAP-1) (Guanylate kinase-associated protein) (SAP90/PSD-95-associated protein 1) (SAPAP1) (PSD-95/SAP90- binding protein 1). | |||||
|
DLGP1_HUMAN
|
||||||
| NC score | 0.961370 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O14490, O14489, P78335 | Gene names | DLGAP1, DAP1, GKAP | |||
|
Domain Architecture |

|
|||||
| Description | Disks large-associated protein 1 (DAP-1) (Guanylate kinase-associated protein) (hGKAP) (SAP90/PSD-95-associated protein 1) (SAPAP1) (PSD- 95/SAP90-binding protein 1). | |||||
|
DLGP2_MOUSE
|
||||||
| NC score | 0.948310 (rank : 4) | θ value | 1.74296e-136 (rank : 4) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BJ42, Q6XBF3 | Gene names | Dlgap2, Dap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 2 (DAP-2) (SAP90/PSD-95-associated protein 2) (SAPAP2) (PSD-95/SAP90-binding protein 2). | |||||
|
DLGP3_MOUSE
|
||||||
| NC score | 0.883220 (rank : 5) | θ value | 2.25659e-51 (rank : 5) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PFD5, Q6PDX0, Q6XBF2 | Gene names | Dlgap3, Dap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
DLGP3_HUMAN
|
||||||
| NC score | 0.882698 (rank : 6) | θ value | 3.84914e-51 (rank : 6) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95886, Q5TDD5, Q9H3X7 | Gene names | DLGAP3, DAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
DLG7_MOUSE
|
||||||
| NC score | 0.610592 (rank : 7) | θ value | 1.13037e-18 (rank : 7) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K4R9, Q6ZQL3, Q80VS7, Q8BUC8, Q8BZ79 | Gene names | Dlg7, Kiaa0008 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discs large homolog 7 (Hepatoma up-regulated protein homolog) (HURP). | |||||
|
DLG7_HUMAN
|
||||||
| NC score | 0.589245 (rank : 8) | θ value | 2.60593e-15 (rank : 8) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15398, Q8NG58 | Gene names | DLG7, KIAA0008 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 7 (Hepatoma up-regulated protein) (HURP). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.064559 (rank : 9) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
MUCEN_MOUSE
|
||||||
| NC score | 0.059695 (rank : 10) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R0H2, Q78KL2, Q9DCN9, Q9ULC1, Q9Z2I1 | Gene names | Emcn, Muc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endomucin precursor (Endomucin-1/2). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.058681 (rank : 11) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.040746 (rank : 12) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
FBX41_MOUSE
|
||||||
| NC score | 0.038159 (rank : 13) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6NS60, Q6P7W4, Q6ZPG1 | Gene names | Fbxo41, D6Ertd538e, Kiaa1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.037911 (rank : 14) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
FBX41_HUMAN
|
||||||
| NC score | 0.035354 (rank : 15) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.034355 (rank : 16) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
DPOLZ_MOUSE
|
||||||
| NC score | 0.031610 (rank : 17) | θ value | 0.279714 (rank : 15) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61493, Q9JMD6, Q9QWX6 | Gene names | Rev3l, Polz, Sez4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (Seizure-related protein 4). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.029966 (rank : 18) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
INAR2_MOUSE
|
||||||
| NC score | 0.029225 (rank : 19) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35664, O35238, O35663, O35983 | Gene names | Ifnar2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-alpha/beta receptor beta chain precursor (IFN-alpha-REC) (Type I interferon receptor) (IFN-R) (Interferon alpha/beta receptor 2). | |||||
|
DNL1_HUMAN
|
||||||
| NC score | 0.028905 (rank : 20) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.028401 (rank : 21) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
CARD6_HUMAN
|
||||||
| NC score | 0.028145 (rank : 22) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BX69 | Gene names | CARD6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 6. | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.027955 (rank : 23) | θ value | 0.0563607 (rank : 9) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.026366 (rank : 24) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
SAMN1_MOUSE
|
||||||
| NC score | 0.025510 (rank : 25) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57725, Q8C6S2 | Gene names | Samsn1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM domain-containing protein SAMSN-1 (SAM domain, SH3 domain and nuclear localization signals protein 1). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.025419 (rank : 26) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.024935 (rank : 27) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
TOPRS_MOUSE
|
||||||
| NC score | 0.024595 (rank : 28) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80Z37, Q3U5B5, Q3UPZ1, Q3USN3, Q3UZ55, Q8BXP2, Q8CFF5, Q8CGC8, Q920L3 | Gene names | Topors | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
SASH1_HUMAN
|
||||||
| NC score | 0.023406 (rank : 29) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O94885, Q8TAI0, Q9H7R7 | Gene names | SASH1, KIAA0790, PEPE1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1 (Proline-glutamate repeat- containing protein). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.022961 (rank : 30) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.022112 (rank : 31) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
PLIN_HUMAN
|
||||||
| NC score | 0.021868 (rank : 32) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60240, Q8N5Y6 | Gene names | PLIN | |||
|
Domain Architecture |

|
|||||
| Description | Perilipin (PERI) (Lipid droplet-associated protein). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.021484 (rank : 33) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
ESPL1_HUMAN
|
||||||
| NC score | 0.021291 (rank : 34) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14674 | Gene names | ESPL1, ESP1, KIAA0165 | |||
|
Domain Architecture |

|
|||||
| Description | Separin (EC 3.4.22.49) (Separase) (Caspase-like protein ESPL1) (Extra spindle poles-like 1 protein). | |||||
|
TROAP_HUMAN
|
||||||
| NC score | 0.020191 (rank : 35) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12815 | Gene names | TROAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophinin-associated protein (Tastin) (Trophinin-assisting protein). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.019968 (rank : 36) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.019570 (rank : 37) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
HAP1_MOUSE
|
||||||
| NC score | 0.019167 (rank : 38) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35668, O35636 | Gene names | Hap1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein 1 (HAP-1). | |||||
|
SAPS1_MOUSE
|
||||||
| NC score | 0.018505 (rank : 39) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
CK024_MOUSE
|
||||||
| NC score | 0.017817 (rank : 40) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.017652 (rank : 41) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.017215 (rank : 42) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
ITA6_HUMAN
|
||||||
| NC score | 0.017088 (rank : 43) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23229, Q08443, Q14646, Q16508, Q9UN03 | Gene names | ITGA6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-6 precursor (VLA-6) (CD49f antigen) [Contains: Integrin alpha-6 heavy chain; Integrin alpha-6 light chain]. | |||||
|
IKBE_MOUSE
|
||||||
| NC score | 0.016859 (rank : 44) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54910, Q3U686, Q9CZZ9, Q9D7U3 | Gene names | Nfkbie, Ikbe | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKB-E). | |||||
|
PHLB2_HUMAN
|
||||||
| NC score | 0.016629 (rank : 45) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
AKAP1_MOUSE
|
||||||
| NC score | 0.016072 (rank : 46) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
RTP3_MOUSE
|
||||||
| NC score | 0.015880 (rank : 47) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.015740 (rank : 48) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RPGF1_HUMAN
|
||||||
| NC score | 0.015710 (rank : 49) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
CO002_HUMAN
|
||||||
| NC score | 0.015263 (rank : 50) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
VPS53_MOUSE
|
||||||
| NC score | 0.014884 (rank : 51) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CCB4, Q8C7U0, Q8CIA1, Q9CX82, Q9D6T7 | Gene names | Vps53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associated protein 53. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.014167 (rank : 52) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.013939 (rank : 53) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.013803 (rank : 54) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
REXO4_MOUSE
|
||||||
| NC score | 0.013297 (rank : 55) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PAQ4, Q3UU17, Q8C524 | Gene names | Rexo4, Gm111, Pmc2, Xpmc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 4 (EC 3.1.-.-) (Exonuclease XPMC2) (Prevents mitotic catastrophe 2 protein homolog). | |||||
|
SIPA1_HUMAN
|
||||||
| NC score | 0.012965 (rank : 56) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96FS4, O14518, O60484, O60618 | Gene names | SIPA1, SPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1) (p130 SPA-1). | |||||
|
CTGE5_HUMAN
|
||||||
| NC score | 0.012188 (rank : 57) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
NAGS_MOUSE
|
||||||
| NC score | 0.012087 (rank : 58) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R4H7, Q8C6G6, Q8CI77, Q8K1R8 | Gene names | Nags | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglutamate synthase, mitochondrial precursor (EC 2.3.1.1) (Amino-acid acetyltransferase) [Contains: N-acetylglutamate synthase long form; N-acetylglutamate synthase short form; N-acetylglutamate synthase conserved domain form]. | |||||
|
CTGE6_HUMAN
|
||||||
| NC score | 0.012011 (rank : 59) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86UF2 | Gene names | CTAGE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-6 (cTAGE family member 6). | |||||
|
TACC1_MOUSE
|
||||||
| NC score | 0.011790 (rank : 60) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
E2F3_HUMAN
|
||||||
| NC score | 0.011771 (rank : 61) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00716, Q15000, Q9BZ44 | Gene names | E2F3, KIAA0075 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
PDLI5_MOUSE
|
||||||
| NC score | 0.010716 (rank : 62) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CI51, Q9QYN0, Q9QYN1, Q9QYN2 | Gene names | Pdlim5, Enh | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
IDD_MOUSE
|
||||||
| NC score | 0.010703 (rank : 63) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98154, Q61844 | Gene names | Dgcr2, Dgsc, Idd, Sez-12, Sez12 | |||
|
Domain Architecture |

|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor (Seizure-related membrane-bound adhesion protein). | |||||
|
BRWD1_MOUSE
|
||||||
| NC score | 0.010676 (rank : 64) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
PJA1_HUMAN
|
||||||
| NC score | 0.010327 (rank : 65) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NG27, Q8NG28, Q9HAC1 | Gene names | PJA1, RNF70 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin protein ligase Praja1 (EC 6.3.2.-) (RING finger protein 70). | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.010154 (rank : 66) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
PHF12_MOUSE
|
||||||
| NC score | 0.009881 (rank : 67) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
SREC2_MOUSE
|
||||||
| NC score | 0.009510 (rank : 68) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
OXR1_HUMAN
|
||||||
| NC score | 0.009506 (rank : 69) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N573, Q3LIB5, Q6ZVK9, Q8N8V0, Q9H266, Q9NWC7 | Gene names | OXR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1. | |||||
|
REPS1_HUMAN
|
||||||
| NC score | 0.009465 (rank : 70) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96D71, Q8NDR7, Q8WU62, Q9BXY9 | Gene names | REPS1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
WDR20_HUMAN
|
||||||
| NC score | 0.009058 (rank : 71) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TBZ3, Q8WXX2, Q9UF86 | Gene names | WDR20 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 20 (DMR protein). | |||||
|
NID2_HUMAN
|
||||||
| NC score | 0.008878 (rank : 72) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
QSCN6_MOUSE
|
||||||
| NC score | 0.008530 (rank : 73) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BND5, Q3TDY9, Q3TE19, Q3TR29, Q3UEL4, Q8K041, Q9DBL6 | Gene names | Qscn6, Sox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfhydryl oxidase 1 precursor (EC 1.8.3.2) (Quiescin Q6) (Skin sulfhydryl oxidase) (mSOx). | |||||
|
GASP2_HUMAN
|
||||||
| NC score | 0.008493 (rank : 74) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96D09, Q8NAB4 | Gene names | GPRASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
SYNJ2_HUMAN
|
||||||
| NC score | 0.008022 (rank : 75) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15056, Q5TA13, Q5TA16, Q5TA19, Q86XK0, Q8IZA8, Q9H226 | Gene names | SYNJ2, KIAA0348 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptojanin-2 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 2). | |||||
|
ZF106_HUMAN
|
||||||
| NC score | 0.007362 (rank : 76) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H2Y7, Q6NSD9, Q6PEK1, Q86T43, Q86T45, Q86T50, Q86T58, Q86TA9, Q96M37, Q9H7B8 | Gene names | ZFP106, ZNF474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 homolog (Zfp-106) (Zinc finger protein 474). | |||||
|
ARHG1_HUMAN
|
||||||
| NC score | 0.007329 (rank : 77) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92888, O00513, Q8N4J4, Q96BF4, Q96F17, Q9BSB1 | Gene names | ARHGEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (p115-RhoGEF) (p115RhoGEF) (115 kDa guanine nucleotide exchange factor) (Sub1.5). | |||||
|
TIAM1_HUMAN
|
||||||
| NC score | 0.007303 (rank : 78) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.007167 (rank : 79) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
TF7L2_HUMAN
|
||||||
| NC score | 0.005621 (rank : 80) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQB0, O00185, Q9NQB1, Q9NQB2, Q9NQB3, Q9NQB4, Q9NQB5, Q9NQB6, Q9NQB7, Q9ULC2 | Gene names | TCF7L2, TCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7-like 2 (HMG box transcription factor 4) (T- cell-specific transcription factor 4) (TCF-4) (hTCF-4). | |||||
|
ZN509_MOUSE
|
||||||
| NC score | 0.004879 (rank : 81) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.004878 (rank : 82) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
CCD39_MOUSE
|
||||||
| NC score | 0.004651 (rank : 83) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
ZN174_HUMAN
|
||||||
| NC score | 0.004381 (rank : 84) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15697, Q9BQ34 | Gene names | ZNF174, ZSCAN8 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 174 (AW-1) (Zinc finger and SCAN domain-containing protein 8). | |||||
|
EVI1_HUMAN
|
||||||
| NC score | 0.003960 (rank : 85) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
MK07_MOUSE
|
||||||
| NC score | 0.003622 (rank : 86) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.003584 (rank : 87) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
ACHA2_HUMAN
|
||||||
| NC score | 0.003561 (rank : 88) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15822, Q9HAQ3 | Gene names | CHRNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal acetylcholine receptor protein subunit alpha-2 precursor. | |||||
|
FBLN2_MOUSE
|
||||||
| NC score | 0.003551 (rank : 89) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
ZFP95_HUMAN
|
||||||
| NC score | 0.001575 (rank : 90) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 90 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2L8 | Gene names | ZFP95, KIAA1015 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 95 homolog (Zfp-95). | |||||