Please be patient as the page loads
|
CRBB1_HUMAN
|
||||||
| SwissProt Accessions | P53674 | Gene names | CRYBB1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta crystallin B1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CRBB1_HUMAN
|
||||||
| θ value | 1.13501e-119 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P53674 | Gene names | CRYBB1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta crystallin B1. | |||||
|
CRBB1_MOUSE
|
||||||
| θ value | 4.95767e-99 (rank : 2) | NC score | 0.998578 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WVJ5 | Gene names | Crybb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta crystallin B1 [Contains: Beta crystallin B1B]. | |||||
|
CRBB3_MOUSE
|
||||||
| θ value | 1.27544e-62 (rank : 3) | NC score | 0.991255 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JJU9 | Gene names | Crybb3 | |||
|
Domain Architecture |
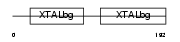
|
|||||
| Description | Beta crystallin B3 (Beta-B3-crystallin). | |||||
|
CRBB3_HUMAN
|
||||||
| θ value | 5.35859e-61 (rank : 4) | NC score | 0.991221 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P26998, Q92965, Q9UH09 | Gene names | CRYBB3, CRYB3 | |||
|
Domain Architecture |
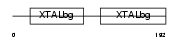
|
|||||
| Description | Beta crystallin B3 (Beta-B3-crystallin). | |||||
|
CRBB2_MOUSE
|
||||||
| θ value | 4.39379e-55 (rank : 5) | NC score | 0.988151 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P62696, P19942, P26775 | Gene names | Crybb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta crystallin B2 (Beta-crystallin Bp). | |||||
|
CRBB2_HUMAN
|
||||||
| θ value | 4.85792e-54 (rank : 6) | NC score | 0.987971 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P43320, Q9UCM8 | Gene names | CRYBB2, CRYB2, CRYB2A | |||
|
Domain Architecture |
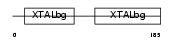
|
|||||
| Description | Beta crystallin B2 (Beta-crystallin Bp). | |||||
|
CRBA1_HUMAN
|
||||||
| θ value | 1.51363e-47 (rank : 7) | NC score | 0.975195 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P05813, Q13633 | Gene names | CRYBA1, CRYB1 | |||
|
Domain Architecture |
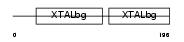
|
|||||
| Description | Beta crystallin A3 [Contains: Beta crystallin A3, isoform A1, Delta4 form; Beta crystallin A3, isoform A1, Delta7 form; Beta crystallin A3, isoform A1, Delta8 form]. | |||||
|
CRBA1_MOUSE
|
||||||
| θ value | 1.08474e-45 (rank : 8) | NC score | 0.973482 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P02525 | Gene names | Cryba1 | |||
|
Domain Architecture |
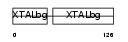
|
|||||
| Description | Beta crystallin A1 (Beta-A1-crystallin). | |||||
|
CRBA4_HUMAN
|
||||||
| θ value | 1.05066e-40 (rank : 9) | NC score | 0.967733 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P53673, Q4VB22, Q6ICE4 | Gene names | CRYBA4 | |||
|
Domain Architecture |
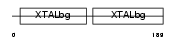
|
|||||
| Description | Beta crystallin A4 (Beta-A4-crystallin). | |||||
|
CRBA4_MOUSE
|
||||||
| θ value | 1.3722e-40 (rank : 10) | NC score | 0.966805 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JJV0 | Gene names | Cryba4 | |||
|
Domain Architecture |
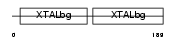
|
|||||
| Description | Beta crystallin A4 (Beta-A4-crystallin). | |||||
|
CRBA2_MOUSE
|
||||||
| θ value | 5.96599e-36 (rank : 11) | NC score | 0.963815 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JJV1 | Gene names | Cryba2 | |||
|
Domain Architecture |
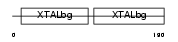
|
|||||
| Description | Beta crystallin A2 (Beta-A2-crystallin). | |||||
|
CRBA2_HUMAN
|
||||||
| θ value | 6.17384e-33 (rank : 12) | NC score | 0.960680 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P53672, Q9Y562 | Gene names | CRYBA2 | |||
|
Domain Architecture |
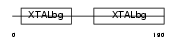
|
|||||
| Description | Beta crystallin A2 (Beta-A2-crystallin). | |||||
|
CRGB_MOUSE
|
||||||
| θ value | 8.63488e-27 (rank : 13) | NC score | 0.904618 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P04344, Q61593 | Gene names | Crygb | |||
|
Domain Architecture |
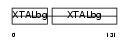
|
|||||
| Description | Gamma crystallin B (Gamma crystallin 3). | |||||
|
CRBS_HUMAN
|
||||||
| θ value | 1.12775e-26 (rank : 14) | NC score | 0.920482 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P22914 | Gene names | CRYGS, GRYG8 | |||
|
Domain Architecture |
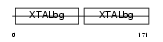
|
|||||
| Description | Beta crystallin S (Gamma crystallin S). | |||||
|
CRBS_MOUSE
|
||||||
| θ value | 1.47289e-26 (rank : 15) | NC score | 0.919423 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35486 | Gene names | Crygs | |||
|
Domain Architecture |
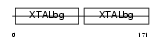
|
|||||
| Description | Beta crystallin S (Gamma crystallin S). | |||||
|
CRGD_MOUSE
|
||||||
| θ value | 5.59698e-26 (rank : 16) | NC score | 0.892579 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P04342, O89027 | Gene names | Crygd | |||
|
Domain Architecture |
|
|||||
| Description | Gamma crystallin D (Gamma crystallin 1). | |||||
|
CRGA_MOUSE
|
||||||
| θ value | 7.30988e-26 (rank : 17) | NC score | 0.900618 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P04345 | Gene names | Cryga | |||
|
Domain Architecture |
|
|||||
| Description | Gamma crystallin A (Gamma crystallin 4). | |||||
|
CRGE_MOUSE
|
||||||
| θ value | 9.54697e-26 (rank : 18) | NC score | 0.892309 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q03740, O89028, P26999, Q9CXK5 | Gene names | Cryge | |||
|
Domain Architecture |
|
|||||
| Description | Gamma crystallin E. | |||||
|
CRGF_MOUSE
|
||||||
| θ value | 4.73814e-25 (rank : 19) | NC score | 0.890901 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9CXV3 | Gene names | Crygf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma crystallin F. | |||||
|
CRGC_MOUSE
|
||||||
| θ value | 1.05554e-24 (rank : 20) | NC score | 0.895232 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61597, Q03739 | Gene names | Crygc, Gammab-cry | |||
|
Domain Architecture |
|
|||||
| Description | Gamma crystallin C. | |||||
|
CRGC_HUMAN
|
||||||
| θ value | 1.37858e-24 (rank : 21) | NC score | 0.902355 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P07315 | Gene names | CRYGC, CRYG3 | |||
|
Domain Architecture |
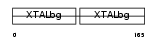
|
|||||
| Description | Gamma crystallin C (Gamma crystallin 2-1) (Gamma crystallin 3). | |||||
|
CRGB_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 22) | NC score | 0.896958 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P07316 | Gene names | CRYGB, CRYG2 | |||
|
Domain Architecture |
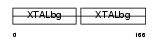
|
|||||
| Description | Gamma crystallin B (Gamma crystallin 1-2). | |||||
|
CRGA_HUMAN
|
||||||
| θ value | 1.6852e-22 (rank : 23) | NC score | 0.895439 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P11844 | Gene names | CRYGA, CRYG1 | |||
|
Domain Architecture |
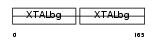
|
|||||
| Description | Gamma crystallin A (Gamma crystallin 5). | |||||
|
CRGD_HUMAN
|
||||||
| θ value | 4.9032e-22 (rank : 24) | NC score | 0.887749 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P07320, Q53R51, Q99681 | Gene names | CRYGD, CRYG4 | |||
|
Domain Architecture |
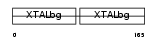
|
|||||
| Description | Gamma crystallin D (Gamma crystallin 4). | |||||
|
AIM1_HUMAN
|
||||||
| θ value | 2.27762e-19 (rank : 25) | NC score | 0.805079 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y4K1, O00296, Q5VWJ2 | Gene names | AIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Absent in melanoma 1 protein. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.007914 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MUCDL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.006569 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
GGA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.009532 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.009283 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SO1C1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.003365 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ERB5, Q8BX54 | Gene names | Slco1c1, Oatpf, Slc21a14 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 1C1 (Solute carrier family 21 member 14) (Organic anion transporter F) (OATP-F) (Organic anion-transporting polypeptide 14) (Organic anion transporter 2) (OATP2). | |||||
|
GGA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.008317 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
CRBB1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.13501e-119 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P53674 | Gene names | CRYBB1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta crystallin B1. | |||||
|
CRBB1_MOUSE
|
||||||
| NC score | 0.998578 (rank : 2) | θ value | 4.95767e-99 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WVJ5 | Gene names | Crybb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta crystallin B1 [Contains: Beta crystallin B1B]. | |||||
|
CRBB3_MOUSE
|
||||||
| NC score | 0.991255 (rank : 3) | θ value | 1.27544e-62 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JJU9 | Gene names | Crybb3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta crystallin B3 (Beta-B3-crystallin). | |||||
|
CRBB3_HUMAN
|
||||||
| NC score | 0.991221 (rank : 4) | θ value | 5.35859e-61 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P26998, Q92965, Q9UH09 | Gene names | CRYBB3, CRYB3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta crystallin B3 (Beta-B3-crystallin). | |||||
|
CRBB2_MOUSE
|
||||||
| NC score | 0.988151 (rank : 5) | θ value | 4.39379e-55 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P62696, P19942, P26775 | Gene names | Crybb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta crystallin B2 (Beta-crystallin Bp). | |||||
|
CRBB2_HUMAN
|
||||||
| NC score | 0.987971 (rank : 6) | θ value | 4.85792e-54 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P43320, Q9UCM8 | Gene names | CRYBB2, CRYB2, CRYB2A | |||
|
Domain Architecture |

|
|||||
| Description | Beta crystallin B2 (Beta-crystallin Bp). | |||||
|
CRBA1_HUMAN
|
||||||
| NC score | 0.975195 (rank : 7) | θ value | 1.51363e-47 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P05813, Q13633 | Gene names | CRYBA1, CRYB1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta crystallin A3 [Contains: Beta crystallin A3, isoform A1, Delta4 form; Beta crystallin A3, isoform A1, Delta7 form; Beta crystallin A3, isoform A1, Delta8 form]. | |||||
|
CRBA1_MOUSE
|
||||||
| NC score | 0.973482 (rank : 8) | θ value | 1.08474e-45 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P02525 | Gene names | Cryba1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta crystallin A1 (Beta-A1-crystallin). | |||||
|
CRBA4_HUMAN
|
||||||
| NC score | 0.967733 (rank : 9) | θ value | 1.05066e-40 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P53673, Q4VB22, Q6ICE4 | Gene names | CRYBA4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta crystallin A4 (Beta-A4-crystallin). | |||||
|
CRBA4_MOUSE
|
||||||
| NC score | 0.966805 (rank : 10) | θ value | 1.3722e-40 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JJV0 | Gene names | Cryba4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta crystallin A4 (Beta-A4-crystallin). | |||||
|
CRBA2_MOUSE
|
||||||
| NC score | 0.963815 (rank : 11) | θ value | 5.96599e-36 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JJV1 | Gene names | Cryba2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta crystallin A2 (Beta-A2-crystallin). | |||||
|
CRBA2_HUMAN
|
||||||
| NC score | 0.960680 (rank : 12) | θ value | 6.17384e-33 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P53672, Q9Y562 | Gene names | CRYBA2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta crystallin A2 (Beta-A2-crystallin). | |||||
|
CRBS_HUMAN
|
||||||
| NC score | 0.920482 (rank : 13) | θ value | 1.12775e-26 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P22914 | Gene names | CRYGS, GRYG8 | |||
|
Domain Architecture |

|
|||||
| Description | Beta crystallin S (Gamma crystallin S). | |||||
|
CRBS_MOUSE
|
||||||
| NC score | 0.919423 (rank : 14) | θ value | 1.47289e-26 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35486 | Gene names | Crygs | |||
|
Domain Architecture |

|
|||||
| Description | Beta crystallin S (Gamma crystallin S). | |||||
|
CRGB_MOUSE
|
||||||
| NC score | 0.904618 (rank : 15) | θ value | 8.63488e-27 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P04344, Q61593 | Gene names | Crygb | |||
|
Domain Architecture |

|
|||||
| Description | Gamma crystallin B (Gamma crystallin 3). | |||||
|
CRGC_HUMAN
|
||||||
| NC score | 0.902355 (rank : 16) | θ value | 1.37858e-24 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P07315 | Gene names | CRYGC, CRYG3 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma crystallin C (Gamma crystallin 2-1) (Gamma crystallin 3). | |||||
|
CRGA_MOUSE
|
||||||
| NC score | 0.900618 (rank : 17) | θ value | 7.30988e-26 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P04345 | Gene names | Cryga | |||
|
Domain Architecture |

|
|||||
| Description | Gamma crystallin A (Gamma crystallin 4). | |||||
|
CRGB_HUMAN
|
||||||
| NC score | 0.896958 (rank : 18) | θ value | 4.43474e-23 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P07316 | Gene names | CRYGB, CRYG2 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma crystallin B (Gamma crystallin 1-2). | |||||
|
CRGA_HUMAN
|
||||||
| NC score | 0.895439 (rank : 19) | θ value | 1.6852e-22 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P11844 | Gene names | CRYGA, CRYG1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma crystallin A (Gamma crystallin 5). | |||||
|
CRGC_MOUSE
|
||||||
| NC score | 0.895232 (rank : 20) | θ value | 1.05554e-24 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61597, Q03739 | Gene names | Crygc, Gammab-cry | |||
|
Domain Architecture |

|
|||||
| Description | Gamma crystallin C. | |||||
|
CRGD_MOUSE
|
||||||
| NC score | 0.892579 (rank : 21) | θ value | 5.59698e-26 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P04342, O89027 | Gene names | Crygd | |||
|
Domain Architecture |

|
|||||
| Description | Gamma crystallin D (Gamma crystallin 1). | |||||
|
CRGE_MOUSE
|
||||||
| NC score | 0.892309 (rank : 22) | θ value | 9.54697e-26 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q03740, O89028, P26999, Q9CXK5 | Gene names | Cryge | |||
|
Domain Architecture |

|
|||||
| Description | Gamma crystallin E. | |||||
|
CRGF_MOUSE
|
||||||
| NC score | 0.890901 (rank : 23) | θ value | 4.73814e-25 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9CXV3 | Gene names | Crygf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma crystallin F. | |||||
|
CRGD_HUMAN
|
||||||
| NC score | 0.887749 (rank : 24) | θ value | 4.9032e-22 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P07320, Q53R51, Q99681 | Gene names | CRYGD, CRYG4 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma crystallin D (Gamma crystallin 4). | |||||
|
AIM1_HUMAN
|
||||||
| NC score | 0.805079 (rank : 25) | θ value | 2.27762e-19 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y4K1, O00296, Q5VWJ2 | Gene names | AIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Absent in melanoma 1 protein. | |||||
|
GGA2_MOUSE
|
||||||
| NC score | 0.009532 (rank : 26) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.009283 (rank : 27) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
GGA2_HUMAN
|
||||||
| NC score | 0.008317 (rank : 28) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.007914 (rank : 29) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MUCDL_MOUSE
|
||||||
| NC score | 0.006569 (rank : 30) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
SO1C1_MOUSE
|
||||||
| NC score | 0.003365 (rank : 31) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ERB5, Q8BX54 | Gene names | Slco1c1, Oatpf, Slc21a14 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 1C1 (Solute carrier family 21 member 14) (Organic anion transporter F) (OATP-F) (Organic anion-transporting polypeptide 14) (Organic anion transporter 2) (OATP2). | |||||