Please be patient as the page loads
|
XDH_MOUSE
|
||||||
| SwissProt Accessions | Q00519 | Gene names | Xdh | |||
|
Domain Architecture |

|
|||||
| Description | Xanthine dehydrogenase/oxidase [Includes: Xanthine dehydrogenase (EC 1.17.1.4) (XD); Xanthine oxidase (EC 1.17.3.2) (XO) (Xanthine oxidoreductase)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ADO_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.985240 (rank : 3) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06278, O14765, Q9UPG6 | Gene names | AOX1, AO | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde oxidase (EC 1.2.3.1). | |||||
|
ADO_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.985073 (rank : 4) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O54754, Q9WU85, Q9Z2Z5 | Gene names | Aox1, Ao, Ro | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde oxidase (EC 1.2.3.1) (Retinal oxidase). | |||||
|
XDH_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999200 (rank : 2) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P47989, Q16681, Q16712, Q4PJ16 | Gene names | XDH, XDHA | |||
|
Domain Architecture |

|
|||||
| Description | Xanthine dehydrogenase/oxidase [Includes: Xanthine dehydrogenase (EC 1.17.1.4) (XD); Xanthine oxidase (EC 1.17.3.2) (XO) (Xanthine oxidoreductase)]. | |||||
|
XDH_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q00519 | Gene names | Xdh | |||
|
Domain Architecture |

|
|||||
| Description | Xanthine dehydrogenase/oxidase [Includes: Xanthine dehydrogenase (EC 1.17.1.4) (XD); Xanthine oxidase (EC 1.17.3.2) (XO) (Xanthine oxidoreductase)]. | |||||
|
KPCE_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 5) | NC score | 0.012585 (rank : 8) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02156, Q53SL4, Q53SM5, Q9UE81 | Gene names | PRKCE, PKCE | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C epsilon type (EC 2.7.11.13) (nPKC-epsilon). | |||||
|
KPCE_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 6) | NC score | 0.012560 (rank : 9) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P16054 | Gene names | Prkce, Pkce, Pkcea | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C epsilon type (EC 2.7.11.13) (nPKC-epsilon). | |||||
|
NEUR1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 7) | NC score | 0.034197 (rank : 6) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99519 | Gene names | NEU1, NANH | |||
|
Domain Architecture |
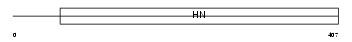
|
|||||
| Description | Sialidase-1 precursor (EC 3.2.1.18) (Lysosomal sialidase) (N-acetyl- alpha-neuraminidase 1) (Acetylneuraminyl hydrolase) (G9 sialidase). | |||||
|
NEUR1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 8) | NC score | 0.034713 (rank : 5) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35657, O55220, Q99KG9 | Gene names | Neu1, Neu | |||
|
Domain Architecture |
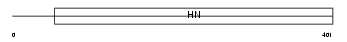
|
|||||
| Description | Sialidase-1 precursor (EC 3.2.1.18) (Lysosomal sialidase) (N-acetyl- alpha-neuraminidase 1) (G9 sialidase). | |||||
|
CD166_MOUSE
|
||||||
| θ value | 3.0926 (rank : 9) | NC score | 0.011134 (rank : 10) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61490, O70136, Q8CDA5, Q8R2T0 | Gene names | Alcam | |||
|
Domain Architecture |
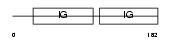
|
|||||
| Description | CD166 antigen precursor (Activated leukocyte-cell adhesion molecule) (ALCAM) (DM-GRASP protein). | |||||
|
MTERF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 10) | NC score | 0.014677 (rank : 7) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CHZ9, Q63ZX5, Q8C7T1 | Gene names | Mterf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor, mitochondrial precursor (mTERF) (Mitochondrial transcription termination factor 1). | |||||
|
TMOD4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 11) | NC score | 0.007753 (rank : 11) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLH8 | Gene names | Tmod4 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-4 (Skeletal muscle tropomodulin) (Sk-Tmod). | |||||
|
XDH_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q00519 | Gene names | Xdh | |||
|
Domain Architecture |

|
|||||
| Description | Xanthine dehydrogenase/oxidase [Includes: Xanthine dehydrogenase (EC 1.17.1.4) (XD); Xanthine oxidase (EC 1.17.3.2) (XO) (Xanthine oxidoreductase)]. | |||||
|
XDH_HUMAN
|
||||||
| NC score | 0.999200 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P47989, Q16681, Q16712, Q4PJ16 | Gene names | XDH, XDHA | |||
|
Domain Architecture |

|
|||||
| Description | Xanthine dehydrogenase/oxidase [Includes: Xanthine dehydrogenase (EC 1.17.1.4) (XD); Xanthine oxidase (EC 1.17.3.2) (XO) (Xanthine oxidoreductase)]. | |||||
|
ADO_HUMAN
|
||||||
| NC score | 0.985240 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06278, O14765, Q9UPG6 | Gene names | AOX1, AO | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde oxidase (EC 1.2.3.1). | |||||
|
ADO_MOUSE
|
||||||
| NC score | 0.985073 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O54754, Q9WU85, Q9Z2Z5 | Gene names | Aox1, Ao, Ro | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde oxidase (EC 1.2.3.1) (Retinal oxidase). | |||||
|
NEUR1_MOUSE
|
||||||
| NC score | 0.034713 (rank : 5) | θ value | 1.81305 (rank : 8) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35657, O55220, Q99KG9 | Gene names | Neu1, Neu | |||
|
Domain Architecture |

|
|||||
| Description | Sialidase-1 precursor (EC 3.2.1.18) (Lysosomal sialidase) (N-acetyl- alpha-neuraminidase 1) (G9 sialidase). | |||||
|
NEUR1_HUMAN
|
||||||
| NC score | 0.034197 (rank : 6) | θ value | 1.81305 (rank : 7) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99519 | Gene names | NEU1, NANH | |||
|
Domain Architecture |

|
|||||
| Description | Sialidase-1 precursor (EC 3.2.1.18) (Lysosomal sialidase) (N-acetyl- alpha-neuraminidase 1) (Acetylneuraminyl hydrolase) (G9 sialidase). | |||||
|
MTERF_MOUSE
|
||||||
| NC score | 0.014677 (rank : 7) | θ value | 6.88961 (rank : 10) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CHZ9, Q63ZX5, Q8C7T1 | Gene names | Mterf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor, mitochondrial precursor (mTERF) (Mitochondrial transcription termination factor 1). | |||||
|
KPCE_HUMAN
|
||||||
| NC score | 0.012585 (rank : 8) | θ value | 0.0736092 (rank : 5) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02156, Q53SL4, Q53SM5, Q9UE81 | Gene names | PRKCE, PKCE | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C epsilon type (EC 2.7.11.13) (nPKC-epsilon). | |||||
|
KPCE_MOUSE
|
||||||
| NC score | 0.012560 (rank : 9) | θ value | 0.0736092 (rank : 6) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P16054 | Gene names | Prkce, Pkce, Pkcea | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C epsilon type (EC 2.7.11.13) (nPKC-epsilon). | |||||
|
CD166_MOUSE
|
||||||
| NC score | 0.011134 (rank : 10) | θ value | 3.0926 (rank : 9) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61490, O70136, Q8CDA5, Q8R2T0 | Gene names | Alcam | |||
|
Domain Architecture |

|
|||||
| Description | CD166 antigen precursor (Activated leukocyte-cell adhesion molecule) (ALCAM) (DM-GRASP protein). | |||||
|
TMOD4_MOUSE
|
||||||
| NC score | 0.007753 (rank : 11) | θ value | 6.88961 (rank : 11) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLH8 | Gene names | Tmod4 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-4 (Skeletal muscle tropomodulin) (Sk-Tmod). | |||||