Please be patient as the page loads
|
ADO_MOUSE
|
||||||
| SwissProt Accessions | O54754, Q9WU85, Q9Z2Z5 | Gene names | Aox1, Ao, Ro | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde oxidase (EC 1.2.3.1) (Retinal oxidase). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ADO_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997812 (rank : 2) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06278, O14765, Q9UPG6 | Gene names | AOX1, AO | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde oxidase (EC 1.2.3.1). | |||||
|
ADO_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O54754, Q9WU85, Q9Z2Z5 | Gene names | Aox1, Ao, Ro | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde oxidase (EC 1.2.3.1) (Retinal oxidase). | |||||
|
XDH_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.985362 (rank : 3) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P47989, Q16681, Q16712, Q4PJ16 | Gene names | XDH, XDHA | |||
|
Domain Architecture |

|
|||||
| Description | Xanthine dehydrogenase/oxidase [Includes: Xanthine dehydrogenase (EC 1.17.1.4) (XD); Xanthine oxidase (EC 1.17.3.2) (XO) (Xanthine oxidoreductase)]. | |||||
|
XDH_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.985073 (rank : 4) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00519 | Gene names | Xdh | |||
|
Domain Architecture |

|
|||||
| Description | Xanthine dehydrogenase/oxidase [Includes: Xanthine dehydrogenase (EC 1.17.1.4) (XD); Xanthine oxidase (EC 1.17.3.2) (XO) (Xanthine oxidoreductase)]. | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 0.21417 (rank : 5) | NC score | 0.010091 (rank : 15) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 1.06291 (rank : 6) | NC score | 0.012594 (rank : 11) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
SYMM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 7) | NC score | 0.020355 (rank : 6) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q499X9, Q3T9W7, Q8BWR9, Q8BXY1 | Gene names | Mars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
AAKG2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 8) | NC score | 0.015919 (rank : 8) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WG5, Q6V7V5 | Gene names | Prkag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-2 (AMPK gamma-2 chain) (AMPK gamma2). | |||||
|
1B35_HUMAN
|
||||||
| θ value | 5.27518 (rank : 9) | NC score | 0.004123 (rank : 17) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30685, O62919, P30468, P30469, P30470, P30471, P30472, P30473, P30474, Q9GJM7, Q9TPV2, Q9TQH3, Q9TQH9, Q9TQN4, Q9TQN6 | Gene names | HLA-B, HLAB | |||
|
Domain Architecture |
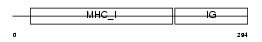
|
|||||
| Description | HLA class I histocompatibility antigen, B-35 alpha chain precursor (MHC class I antigen B*35). | |||||
|
CAND2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 10) | NC score | 0.010459 (rank : 14) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZQ73, Q6PHT7 | Gene names | Cand2, Kiaa0667, Tip120b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2) (TBP-interacting protein TIP120B) (TBP-interacting protein of 120 kDa B). | |||||
|
INP4B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 11) | NC score | 0.011318 (rank : 13) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15327, Q6IN59, Q6PJB4 | Gene names | INPP4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-3,4-bisphosphate 4-phosphatase (EC 3.1.3.66) (Inositol polyphosphate 4-phosphatase type II). | |||||
|
1B45_HUMAN
|
||||||
| θ value | 6.88961 (rank : 12) | NC score | 0.003907 (rank : 19) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30483, Q9GIZ0, Q9MX21 | Gene names | HLA-B, HLAB | |||
|
Domain Architecture |
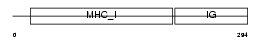
|
|||||
| Description | HLA class I histocompatibility antigen, B-45 alpha chain precursor (MHC class I antigen B*45) (Bw-45). | |||||
|
1B50_HUMAN
|
||||||
| θ value | 6.88961 (rank : 13) | NC score | 0.003900 (rank : 20) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30488, O19615, O19624, O19641, O19783, O46702, O78172, Q9TQG1 | Gene names | HLA-B, HLAB | |||
|
Domain Architecture |
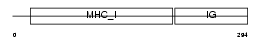
|
|||||
| Description | HLA class I histocompatibility antigen, B-50 alpha chain precursor (MHC class I antigen B*50) (Bw-50) (B-21). | |||||
|
AAKG2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 14) | NC score | 0.014126 (rank : 9) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UGJ0, Q53Y07, Q6NUI0, Q75MP4, Q9NUZ9, Q9UDN8, Q9ULX8 | Gene names | PRKAG2 | |||
|
Domain Architecture |

|
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-2 (AMPK gamma-2 chain) (AMPK gamma2) (H91620p). | |||||
|
I12R1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 15) | NC score | 0.021928 (rank : 5) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60837 | Gene names | Il12rb1, Il12rb | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-12 receptor beta-1 chain precursor (IL-12R-beta1) (Interleukin-12 receptor beta) (IL-12 receptor beta component) (CD212 antigen). | |||||
|
KIF6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 16) | NC score | 0.004450 (rank : 16) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZMV9, Q2MDE3, Q2MDE4, Q5T8J6, Q6ZWE3, Q86T87, Q8WTV4 | Gene names | KIF6, C6orf102 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF6. | |||||
|
LY6H_HUMAN
|
||||||
| θ value | 6.88961 (rank : 17) | NC score | 0.016514 (rank : 7) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94772 | Gene names | LY6H | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen Ly-6H precursor. | |||||
|
MYO9B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.002905 (rank : 23) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
1C15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 19) | NC score | 0.003603 (rank : 22) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07000, O78165, Q29864, Q29991, Q31605, Q9BD28, Q9GJ33, Q9MY49 | Gene names | HLA-C, HLAC | |||
|
Domain Architecture |
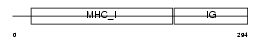
|
|||||
| Description | HLA class I histocompatibility antigen, Cw-15 alpha chain precursor (MHC class I antigen Cw*15). | |||||
|
CAN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.003726 (rank : 21) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07384 | Gene names | CAPN1, CANPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
CI009_HUMAN
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.013251 (rank : 10) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96E40, Q9UGQ0 | Gene names | C9orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf9. | |||||
|
CNTP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.004041 (rank : 18) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9C0A0 | Gene names | CNTNAP4, CASPR4, KIAA1763 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
CR2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.011661 (rank : 12) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19070 | Gene names | Cr2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 2 precursor (Cr2) (Complement C3d receptor) (CD21 antigen). | |||||
|
ADO_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O54754, Q9WU85, Q9Z2Z5 | Gene names | Aox1, Ao, Ro | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde oxidase (EC 1.2.3.1) (Retinal oxidase). | |||||
|
ADO_HUMAN
|
||||||
| NC score | 0.997812 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06278, O14765, Q9UPG6 | Gene names | AOX1, AO | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde oxidase (EC 1.2.3.1). | |||||
|
XDH_HUMAN
|
||||||
| NC score | 0.985362 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P47989, Q16681, Q16712, Q4PJ16 | Gene names | XDH, XDHA | |||
|
Domain Architecture |

|
|||||
| Description | Xanthine dehydrogenase/oxidase [Includes: Xanthine dehydrogenase (EC 1.17.1.4) (XD); Xanthine oxidase (EC 1.17.3.2) (XO) (Xanthine oxidoreductase)]. | |||||
|
XDH_MOUSE
|
||||||
| NC score | 0.985073 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00519 | Gene names | Xdh | |||
|
Domain Architecture |

|
|||||
| Description | Xanthine dehydrogenase/oxidase [Includes: Xanthine dehydrogenase (EC 1.17.1.4) (XD); Xanthine oxidase (EC 1.17.3.2) (XO) (Xanthine oxidoreductase)]. | |||||
|
I12R1_MOUSE
|
||||||
| NC score | 0.021928 (rank : 5) | θ value | 6.88961 (rank : 15) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60837 | Gene names | Il12rb1, Il12rb | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-12 receptor beta-1 chain precursor (IL-12R-beta1) (Interleukin-12 receptor beta) (IL-12 receptor beta component) (CD212 antigen). | |||||
|
SYMM_MOUSE
|
||||||
| NC score | 0.020355 (rank : 6) | θ value | 2.36792 (rank : 7) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q499X9, Q3T9W7, Q8BWR9, Q8BXY1 | Gene names | Mars2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.10) (Methionine--tRNA ligase 2) (Mitochondrial methionine--tRNA ligase) (MtMetRS). | |||||
|
LY6H_HUMAN
|
||||||
| NC score | 0.016514 (rank : 7) | θ value | 6.88961 (rank : 17) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94772 | Gene names | LY6H | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen Ly-6H precursor. | |||||
|
AAKG2_MOUSE
|
||||||
| NC score | 0.015919 (rank : 8) | θ value | 4.03905 (rank : 8) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WG5, Q6V7V5 | Gene names | Prkag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-2 (AMPK gamma-2 chain) (AMPK gamma2). | |||||
|
AAKG2_HUMAN
|
||||||
| NC score | 0.014126 (rank : 9) | θ value | 6.88961 (rank : 14) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UGJ0, Q53Y07, Q6NUI0, Q75MP4, Q9NUZ9, Q9UDN8, Q9ULX8 | Gene names | PRKAG2 | |||
|
Domain Architecture |

|
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-2 (AMPK gamma-2 chain) (AMPK gamma2) (H91620p). | |||||
|
CI009_HUMAN
|
||||||
| NC score | 0.013251 (rank : 10) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96E40, Q9UGQ0 | Gene names | C9orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf9. | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.012594 (rank : 11) | θ value | 1.06291 (rank : 6) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
CR2_MOUSE
|
||||||
| NC score | 0.011661 (rank : 12) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19070 | Gene names | Cr2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement receptor type 2 precursor (Cr2) (Complement C3d receptor) (CD21 antigen). | |||||
|
INP4B_HUMAN
|
||||||
| NC score | 0.011318 (rank : 13) | θ value | 5.27518 (rank : 11) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15327, Q6IN59, Q6PJB4 | Gene names | INPP4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-3,4-bisphosphate 4-phosphatase (EC 3.1.3.66) (Inositol polyphosphate 4-phosphatase type II). | |||||
|
CAND2_MOUSE
|
||||||
| NC score | 0.010459 (rank : 14) | θ value | 5.27518 (rank : 10) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZQ73, Q6PHT7 | Gene names | Cand2, Kiaa0667, Tip120b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2) (TBP-interacting protein TIP120B) (TBP-interacting protein of 120 kDa B). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.010091 (rank : 15) | θ value | 0.21417 (rank : 5) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
KIF6_HUMAN
|
||||||
| NC score | 0.004450 (rank : 16) | θ value | 6.88961 (rank : 16) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZMV9, Q2MDE3, Q2MDE4, Q5T8J6, Q6ZWE3, Q86T87, Q8WTV4 | Gene names | KIF6, C6orf102 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF6. | |||||
|
1B35_HUMAN
|
||||||
| NC score | 0.004123 (rank : 17) | θ value | 5.27518 (rank : 9) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30685, O62919, P30468, P30469, P30470, P30471, P30472, P30473, P30474, Q9GJM7, Q9TPV2, Q9TQH3, Q9TQH9, Q9TQN4, Q9TQN6 | Gene names | HLA-B, HLAB | |||
|
Domain Architecture |

|
|||||
| Description | HLA class I histocompatibility antigen, B-35 alpha chain precursor (MHC class I antigen B*35). | |||||
|
CNTP4_HUMAN
|
||||||
| NC score | 0.004041 (rank : 18) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9C0A0 | Gene names | CNTNAP4, CASPR4, KIAA1763 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
1B45_HUMAN
|
||||||
| NC score | 0.003907 (rank : 19) | θ value | 6.88961 (rank : 12) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30483, Q9GIZ0, Q9MX21 | Gene names | HLA-B, HLAB | |||
|
Domain Architecture |

|
|||||
| Description | HLA class I histocompatibility antigen, B-45 alpha chain precursor (MHC class I antigen B*45) (Bw-45). | |||||
|
1B50_HUMAN
|
||||||
| NC score | 0.003900 (rank : 20) | θ value | 6.88961 (rank : 13) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30488, O19615, O19624, O19641, O19783, O46702, O78172, Q9TQG1 | Gene names | HLA-B, HLAB | |||
|
Domain Architecture |

|
|||||
| Description | HLA class I histocompatibility antigen, B-50 alpha chain precursor (MHC class I antigen B*50) (Bw-50) (B-21). | |||||
|
CAN1_HUMAN
|
||||||
| NC score | 0.003726 (rank : 21) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07384 | Gene names | CAPN1, CANPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-1 catalytic subunit (EC 3.4.22.52) (Calpain-1 large subunit) (Calcium-activated neutral proteinase 1) (CANP 1) (Calpain mu-type) (muCANP) (Micromolar-calpain). | |||||
|
1C15_HUMAN
|
||||||
| NC score | 0.003603 (rank : 22) | θ value | 8.99809 (rank : 19) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07000, O78165, Q29864, Q29991, Q31605, Q9BD28, Q9GJ33, Q9MY49 | Gene names | HLA-C, HLAC | |||
|
Domain Architecture |

|
|||||
| Description | HLA class I histocompatibility antigen, Cw-15 alpha chain precursor (MHC class I antigen Cw*15). | |||||
|
MYO9B_MOUSE
|
||||||
| NC score | 0.002905 (rank : 23) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||