Please be patient as the page loads
|
VAT1_MOUSE
|
||||||
| SwissProt Accessions | Q62465, Q8R3G0, Q8R3T8 | Gene names | Vat1, Vat-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptic vesicle membrane protein VAT-1 homolog (EC 1.-.-.-). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
VAT1_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62465, Q8R3G0, Q8R3T8 | Gene names | Vat1, Vat-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptic vesicle membrane protein VAT-1 homolog (EC 1.-.-.-). | |||||
|
VAT1_HUMAN
|
||||||
| θ value | 3.24919e-175 (rank : 2) | NC score | 0.997867 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99536, Q13035, Q96A39, Q9BUT8 | Gene names | VAT1 | |||
|
Domain Architecture |
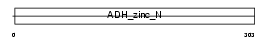
|
|||||
| Description | Synaptic vesicle membrane protein VAT-1 homolog (EC 1.-.-.-). | |||||
|
K1576_MOUSE
|
||||||
| θ value | 8.79181e-80 (rank : 3) | NC score | 0.959031 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80TB8, Q6PGM9 | Gene names | Kiaa1576 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase KIAA1576 (EC 1.-.-.-). | |||||
|
K1576_HUMAN
|
||||||
| θ value | 4.36332e-79 (rank : 4) | NC score | 0.959809 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HCJ6, Q8IYW8 | Gene names | KIAA1576 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase KIAA1576 (EC 1.-.-.-). | |||||
|
QOR_HUMAN
|
||||||
| θ value | 1.6774e-38 (rank : 5) | NC score | 0.881339 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q08257, Q53FT0, Q59EU7, Q6NSK9 | Gene names | CRYZ | |||
|
Domain Architecture |
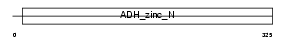
|
|||||
| Description | Quinone oxidoreductase (EC 1.6.5.5) (NADPH:quinone reductase) (Zeta- crystallin). | |||||
|
QOR_MOUSE
|
||||||
| θ value | 2.12192e-33 (rank : 6) | NC score | 0.879711 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P47199, Q62508, Q99L63 | Gene names | Cryz | |||
|
Domain Architecture |
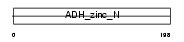
|
|||||
| Description | Quinone oxidoreductase (EC 1.6.5.5) (NADPH:quinone reductase) (Zeta- crystallin). | |||||
|
QORX_HUMAN
|
||||||
| θ value | 8.06329e-33 (rank : 7) | NC score | 0.891641 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q53FA7, O14679, O14685, Q38G78, Q6JLE7, Q9BWB8 | Gene names | TP53I3, PIG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative quinone oxidoreductase (EC 1.-.-.-) (Tumor protein p53- inducible protein 3) (p53-induced protein 3). | |||||
|
ZADH2_HUMAN
|
||||||
| θ value | 8.93572e-24 (rank : 8) | NC score | 0.750089 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N4Q0 | Gene names | ZADH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 2 (EC 1.-.-.-). | |||||
|
ZADH2_MOUSE
|
||||||
| θ value | 3.75424e-22 (rank : 9) | NC score | 0.756604 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BGC4 | Gene names | Zadh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 2 (EC 1.-.-.-). | |||||
|
MECR_MOUSE
|
||||||
| θ value | 3.88503e-19 (rank : 10) | NC score | 0.747259 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9DCS3, Q99L39 | Gene names | Mecr, Nrbf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-2-enoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.38). | |||||
|
MECR_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 11) | NC score | 0.720831 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BV79, Q5SYU0, Q5SYU1, Q5SYU2, Q6IBU9, Q9Y373 | Gene names | MECR, NBRF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-2-enoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.38) (HsNrbf-1) (NRBF-1). | |||||
|
RT4I1_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 12) | NC score | 0.641551 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q924D0, Q8R1T0 | Gene names | Rtn4ip1, Nimp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4-interacting protein 1, mitochondrial precursor (NOGO- interacting mitochondrial protein). | |||||
|
FAS_MOUSE
|
||||||
| θ value | 3.18553e-13 (rank : 13) | NC score | 0.536570 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
RT4I1_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 14) | NC score | 0.640779 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WWV3, Q8N9B3, Q8WZ66, Q9BRA4 | Gene names | RTN4IP1, NIMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4-interacting protein 1, mitochondrial precursor (NOGO- interacting mitochondrial protein). | |||||
|
FAS_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 15) | NC score | 0.522728 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49327, Q13479, Q16702, Q6P4U5, Q6SS02, Q969R1, Q96C68, Q96IT0 | Gene names | FASN, FAS | |||
|
Domain Architecture |

|
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
DHSO_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 16) | NC score | 0.461478 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q00796, Q16682, Q9UMD6 | Gene names | SORD | |||
|
Domain Architecture |
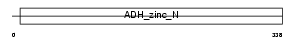
|
|||||
| Description | Sorbitol dehydrogenase (EC 1.1.1.14) (L-iditol 2-dehydrogenase). | |||||
|
ADH1A_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 17) | NC score | 0.286538 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P07327 | Gene names | ADH1A, ADH1 | |||
|
Domain Architecture |
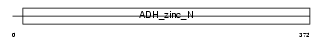
|
|||||
| Description | Alcohol dehydrogenase 1A (EC 1.1.1.1) (Alcohol dehydrogenase alpha subunit). | |||||
|
QORL_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 18) | NC score | 0.486438 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95825, Q96DY0, Q9NVY7 | Gene names | CRYZL1 | |||
|
Domain Architecture |
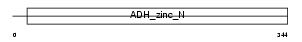
|
|||||
| Description | Quinone oxidoreductase-like 1 (EC 1.-.-.-) (QOH-1) (Zeta-crystallin homolog) (4P11). | |||||
|
ADH1B_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 19) | NC score | 0.286057 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P00325, Q13711, Q4ZGI9, Q96KI7 | Gene names | ADH1B, ADH2 | |||
|
Domain Architecture |
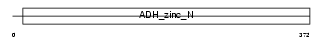
|
|||||
| Description | Alcohol dehydrogenase 1B (EC 1.1.1.1) (Alcohol dehydrogenase beta subunit). | |||||
|
LTB4D_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 20) | NC score | 0.405263 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91YR9, Q3TKT6, Q9CPS1 | Gene names | Ltb4dh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADP-dependent leukotriene B4 12-hydroxydehydrogenase (EC 1.3.1.74) (15-oxoprostaglandin 13-reductase) (EC 1.3.1.48). | |||||
|
QORL_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 21) | NC score | 0.462855 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q921W4, Q3UKX2, Q8VCS1, Q9CWR9 | Gene names | Cryzl1 | |||
|
Domain Architecture |
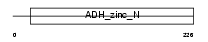
|
|||||
| Description | Quinone oxidoreductase-like 1 (EC 1.-.-.-) (QOH-1) (Zeta-crystallin homolog). | |||||
|
ADH1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 22) | NC score | 0.254206 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P00329 | Gene names | Adh1, Adh-1 | |||
|
Domain Architecture |
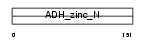
|
|||||
| Description | Alcohol dehydrogenase 1 (EC 1.1.1.1) (Alcohol dehydrogenase A subunit) (ADH-A2). | |||||
|
ADH1G_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 23) | NC score | 0.272192 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P00326, Q4PJ18, Q5WRV0, Q6LBW4, Q6NWV0, Q6NZA7 | Gene names | ADH1C, ADH3 | |||
|
Domain Architecture |
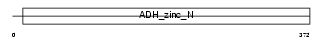
|
|||||
| Description | Alcohol dehydrogenase 1C (EC 1.1.1.1) (Alcohol dehydrogenase gamma subunit). | |||||
|
ADH4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 24) | NC score | 0.235791 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P08319, Q8TCD7 | Gene names | ADH4 | |||
|
Domain Architecture |
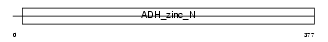
|
|||||
| Description | Alcohol dehydrogenase 4 (EC 1.1.1.1) (Alcohol dehydrogenase class II pi chain). | |||||
|
ADHX_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 25) | NC score | 0.236528 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P28474, Q3TW83, Q8C662 | Gene names | Adh5, Adh-2, Adh2 | |||
|
Domain Architecture |
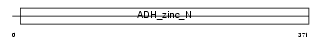
|
|||||
| Description | Alcohol dehydrogenase class 3 (EC 1.1.1.1) (Alcohol dehydrogenase class III) (Alcohol dehydrogenase 2) (S-(hydroxymethyl)glutathione dehydrogenase) (EC 1.1.1.284) (Glutathione-dependent formaldehyde dehydrogenase) (FDH) (FALDH) (Alcohol dehydrogenase-B2) (ADH-B2). | |||||
|
ADHX_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 26) | NC score | 0.246304 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11766, Q6FHR2 | Gene names | ADH5, ADHX, FDH | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase class 3 chi chain (EC 1.1.1.1) (Alcohol dehydrogenase class III chi chain) (S-(hydroxymethyl)glutathione dehydrogenase) (EC 1.1.1.284) (Glutathione-dependent formaldehyde dehydrogenase) (FDH). | |||||
|
ADH6_HUMAN
|
||||||
| θ value | 0.125558 (rank : 27) | NC score | 0.222573 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P28332, Q58F53 | Gene names | ADH6 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 6 (EC 1.1.1.1). | |||||
|
LTB4D_HUMAN
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.376242 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14914, Q8IYQ0, Q9H1X6 | Gene names | LTB4DH | |||
|
Domain Architecture |
|
|||||
| Description | NADP-dependent leukotriene B4 12-hydroxydehydrogenase (EC 1.3.1.74) (15-oxoprostaglandin 13-reductase) (EC 1.3.1.48). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.010343 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.013660 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
BGH3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.026728 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P82198, Q3U9R1 | Gene names | Tgfbi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming growth factor-beta-induced protein ig-h3 precursor (Beta ig-h3). | |||||
|
IL4RA_MOUSE
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.026430 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
ADH7_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.227515 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P40394, Q13713 | Gene names | ADH7 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase class 4 mu/sigma chain (EC 1.1.1.1) (Alcohol dehydrogenase class IV mu/sigma chain) (Retinol dehydrogenase) (Gastric alcohol dehydrogenase). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.016849 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
ADH4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.213435 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QYY9 | Gene names | Adh4 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 4 (EC 1.1.1.1) (Alcohol dehydrogenase class II) (Alcohol dehydrogenase II) (ADH2). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | -0.001482 (rank : 45) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.009220 (rank : 42) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.004834 (rank : 43) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CSPG5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.010385 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | -0.002358 (rank : 46) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MGR3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.002990 (rank : 44) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14832, Q75MV4, Q75N17, Q86YG6, Q8TBH9 | Gene names | GRM3, GPRC1C, MGLUR3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
ADH7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.199184 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64437 | Gene names | Adh7, Adh-3, Adh3 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase class 4 mu/sigma chain (EC 1.1.1.1) (Alcohol dehydrogenase class IV mu/sigma chain) (Retinol dehydrogenase) (Gastric alcohol dehydrogenase) (ADH-C2) (Alcohol dehydrogenase 7). | |||||
|
OXSM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.066149 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NWU1 | Gene names | OXSM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial precursor (EC 2.3.1.41) (Beta-ketoacyl synthase). | |||||
|
OXSM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.067749 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D404 | Gene names | Oxsm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial precursor (EC 2.3.1.41) (Beta-ketoacyl synthase). | |||||
|
ZADH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.141808 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N8N7, Q6MZH8 | Gene names | ZADH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
ZADH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.126074 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VDQ1, Q8BZA2, Q9D1W8 | Gene names | Zadh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
VAT1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62465, Q8R3G0, Q8R3T8 | Gene names | Vat1, Vat-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptic vesicle membrane protein VAT-1 homolog (EC 1.-.-.-). | |||||
|
VAT1_HUMAN
|
||||||
| NC score | 0.997867 (rank : 2) | θ value | 3.24919e-175 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99536, Q13035, Q96A39, Q9BUT8 | Gene names | VAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptic vesicle membrane protein VAT-1 homolog (EC 1.-.-.-). | |||||
|
K1576_HUMAN
|
||||||
| NC score | 0.959809 (rank : 3) | θ value | 4.36332e-79 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HCJ6, Q8IYW8 | Gene names | KIAA1576 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase KIAA1576 (EC 1.-.-.-). | |||||
|
K1576_MOUSE
|
||||||
| NC score | 0.959031 (rank : 4) | θ value | 8.79181e-80 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80TB8, Q6PGM9 | Gene names | Kiaa1576 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase KIAA1576 (EC 1.-.-.-). | |||||
|
QORX_HUMAN
|
||||||
| NC score | 0.891641 (rank : 5) | θ value | 8.06329e-33 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q53FA7, O14679, O14685, Q38G78, Q6JLE7, Q9BWB8 | Gene names | TP53I3, PIG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative quinone oxidoreductase (EC 1.-.-.-) (Tumor protein p53- inducible protein 3) (p53-induced protein 3). | |||||
|
QOR_HUMAN
|
||||||
| NC score | 0.881339 (rank : 6) | θ value | 1.6774e-38 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q08257, Q53FT0, Q59EU7, Q6NSK9 | Gene names | CRYZ | |||
|
Domain Architecture |
|
|||||
| Description | Quinone oxidoreductase (EC 1.6.5.5) (NADPH:quinone reductase) (Zeta- crystallin). | |||||
|
QOR_MOUSE
|
||||||
| NC score | 0.879711 (rank : 7) | θ value | 2.12192e-33 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P47199, Q62508, Q99L63 | Gene names | Cryz | |||
|
Domain Architecture |

|
|||||
| Description | Quinone oxidoreductase (EC 1.6.5.5) (NADPH:quinone reductase) (Zeta- crystallin). | |||||
|
ZADH2_MOUSE
|
||||||
| NC score | 0.756604 (rank : 8) | θ value | 3.75424e-22 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BGC4 | Gene names | Zadh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 2 (EC 1.-.-.-). | |||||
|
ZADH2_HUMAN
|
||||||
| NC score | 0.750089 (rank : 9) | θ value | 8.93572e-24 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N4Q0 | Gene names | ZADH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 2 (EC 1.-.-.-). | |||||
|
MECR_MOUSE
|
||||||
| NC score | 0.747259 (rank : 10) | θ value | 3.88503e-19 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9DCS3, Q99L39 | Gene names | Mecr, Nrbf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-2-enoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.38). | |||||
|
MECR_HUMAN
|
||||||
| NC score | 0.720831 (rank : 11) | θ value | 3.07829e-16 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BV79, Q5SYU0, Q5SYU1, Q5SYU2, Q6IBU9, Q9Y373 | Gene names | MECR, NBRF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-2-enoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.38) (HsNrbf-1) (NRBF-1). | |||||
|
RT4I1_MOUSE
|
||||||
| NC score | 0.641551 (rank : 12) | θ value | 1.09485e-13 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q924D0, Q8R1T0 | Gene names | Rtn4ip1, Nimp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4-interacting protein 1, mitochondrial precursor (NOGO- interacting mitochondrial protein). | |||||
|
RT4I1_HUMAN
|
||||||
| NC score | 0.640779 (rank : 13) | θ value | 9.26847e-13 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WWV3, Q8N9B3, Q8WZ66, Q9BRA4 | Gene names | RTN4IP1, NIMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4-interacting protein 1, mitochondrial precursor (NOGO- interacting mitochondrial protein). | |||||
|
FAS_MOUSE
|
||||||
| NC score | 0.536570 (rank : 14) | θ value | 3.18553e-13 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
FAS_HUMAN
|
||||||
| NC score | 0.522728 (rank : 15) | θ value | 5.08577e-11 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49327, Q13479, Q16702, Q6P4U5, Q6SS02, Q969R1, Q96C68, Q96IT0 | Gene names | FASN, FAS | |||
|
Domain Architecture |

|
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
QORL_HUMAN
|
||||||
| NC score | 0.486438 (rank : 16) | θ value | 0.000158464 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95825, Q96DY0, Q9NVY7 | Gene names | CRYZL1 | |||
|
Domain Architecture |

|
|||||
| Description | Quinone oxidoreductase-like 1 (EC 1.-.-.-) (QOH-1) (Zeta-crystallin homolog) (4P11). | |||||
|
QORL_MOUSE
|
||||||
| NC score | 0.462855 (rank : 17) | θ value | 0.00665767 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q921W4, Q3UKX2, Q8VCS1, Q9CWR9 | Gene names | Cryzl1 | |||
|
Domain Architecture |

|
|||||
| Description | Quinone oxidoreductase-like 1 (EC 1.-.-.-) (QOH-1) (Zeta-crystallin homolog). | |||||
|
DHSO_HUMAN
|
||||||
| NC score | 0.461478 (rank : 18) | θ value | 9.92553e-07 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q00796, Q16682, Q9UMD6 | Gene names | SORD | |||
|
Domain Architecture |
|
|||||
| Description | Sorbitol dehydrogenase (EC 1.1.1.14) (L-iditol 2-dehydrogenase). | |||||
|
LTB4D_MOUSE
|
||||||
| NC score | 0.405263 (rank : 19) | θ value | 0.000461057 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91YR9, Q3TKT6, Q9CPS1 | Gene names | Ltb4dh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADP-dependent leukotriene B4 12-hydroxydehydrogenase (EC 1.3.1.74) (15-oxoprostaglandin 13-reductase) (EC 1.3.1.48). | |||||
|
LTB4D_HUMAN
|
||||||
| NC score | 0.376242 (rank : 20) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14914, Q8IYQ0, Q9H1X6 | Gene names | LTB4DH | |||
|
Domain Architecture |
|
|||||
| Description | NADP-dependent leukotriene B4 12-hydroxydehydrogenase (EC 1.3.1.74) (15-oxoprostaglandin 13-reductase) (EC 1.3.1.48). | |||||
|
ADH1A_HUMAN
|
||||||
| NC score | 0.286538 (rank : 21) | θ value | 5.44631e-05 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P07327 | Gene names | ADH1A, ADH1 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 1A (EC 1.1.1.1) (Alcohol dehydrogenase alpha subunit). | |||||
|
ADH1B_HUMAN
|
||||||
| NC score | 0.286057 (rank : 22) | θ value | 0.00035302 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P00325, Q13711, Q4ZGI9, Q96KI7 | Gene names | ADH1B, ADH2 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 1B (EC 1.1.1.1) (Alcohol dehydrogenase beta subunit). | |||||
|
ADH1G_HUMAN
|
||||||
| NC score | 0.272192 (rank : 23) | θ value | 0.0252991 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P00326, Q4PJ18, Q5WRV0, Q6LBW4, Q6NWV0, Q6NZA7 | Gene names | ADH1C, ADH3 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 1C (EC 1.1.1.1) (Alcohol dehydrogenase gamma subunit). | |||||
|
ADH1_MOUSE
|
||||||
| NC score | 0.254206 (rank : 24) | θ value | 0.0193708 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P00329 | Gene names | Adh1, Adh-1 | |||
|
Domain Architecture |

|
|||||
| Description | Alcohol dehydrogenase 1 (EC 1.1.1.1) (Alcohol dehydrogenase A subunit) (ADH-A2). | |||||
|
ADHX_HUMAN
|
||||||
| NC score | 0.246304 (rank : 25) | θ value | 0.0431538 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11766, Q6FHR2 | Gene names | ADH5, ADHX, FDH | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase class 3 chi chain (EC 1.1.1.1) (Alcohol dehydrogenase class III chi chain) (S-(hydroxymethyl)glutathione dehydrogenase) (EC 1.1.1.284) (Glutathione-dependent formaldehyde dehydrogenase) (FDH). | |||||
|
ADHX_MOUSE
|
||||||
| NC score | 0.236528 (rank : 26) | θ value | 0.0330416 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P28474, Q3TW83, Q8C662 | Gene names | Adh5, Adh-2, Adh2 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase class 3 (EC 1.1.1.1) (Alcohol dehydrogenase class III) (Alcohol dehydrogenase 2) (S-(hydroxymethyl)glutathione dehydrogenase) (EC 1.1.1.284) (Glutathione-dependent formaldehyde dehydrogenase) (FDH) (FALDH) (Alcohol dehydrogenase-B2) (ADH-B2). | |||||
|
ADH4_HUMAN
|
||||||
| NC score | 0.235791 (rank : 27) | θ value | 0.0330416 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P08319, Q8TCD7 | Gene names | ADH4 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase 4 (EC 1.1.1.1) (Alcohol dehydrogenase class II pi chain). | |||||
|
ADH7_HUMAN
|
||||||
| NC score | 0.227515 (rank : 28) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P40394, Q13713 | Gene names | ADH7 | |||
|
Domain Architecture |
|
|||||
| Description | Alcohol dehydrogenase class 4 mu/sigma chain (EC 1.1.1.1) (Alcohol dehydrogenase class IV mu/sigma chain) (Retinol dehydrogenase) (Gastric alcohol dehydrogenase). | |||||
|
ADH6_HUMAN
|
||||||
| NC score | 0.222573 (rank : 29) | θ value | 0.125558 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P28332, Q58F53 | Gene names | ADH6 | |||
|
Domain Architecture |
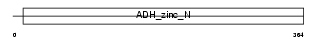
|
|||||
| Description | Alcohol dehydrogenase 6 (EC 1.1.1.1). | |||||
|
ADH4_MOUSE
|
||||||
| NC score | 0.213435 (rank : 30) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QYY9 | Gene names | Adh4 | |||
|
Domain Architecture |
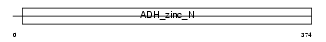
|
|||||
| Description | Alcohol dehydrogenase 4 (EC 1.1.1.1) (Alcohol dehydrogenase class II) (Alcohol dehydrogenase II) (ADH2). | |||||
|
ADH7_MOUSE
|
||||||
| NC score | 0.199184 (rank : 31) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64437 | Gene names | Adh7, Adh-3, Adh3 | |||
|
Domain Architecture |
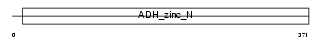
|
|||||
| Description | Alcohol dehydrogenase class 4 mu/sigma chain (EC 1.1.1.1) (Alcohol dehydrogenase class IV mu/sigma chain) (Retinol dehydrogenase) (Gastric alcohol dehydrogenase) (ADH-C2) (Alcohol dehydrogenase 7). | |||||
|
ZADH1_HUMAN
|
||||||
| NC score | 0.141808 (rank : 32) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N8N7, Q6MZH8 | Gene names | ZADH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
ZADH1_MOUSE
|
||||||
| NC score | 0.126074 (rank : 33) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VDQ1, Q8BZA2, Q9D1W8 | Gene names | Zadh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-binding alcohol dehydrogenase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
OXSM_MOUSE
|
||||||
| NC score | 0.067749 (rank : 34) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D404 | Gene names | Oxsm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial precursor (EC 2.3.1.41) (Beta-ketoacyl synthase). | |||||
|
OXSM_HUMAN
|
||||||
| NC score | 0.066149 (rank : 35) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NWU1 | Gene names | OXSM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial precursor (EC 2.3.1.41) (Beta-ketoacyl synthase). | |||||
|
BGH3_MOUSE
|
||||||
| NC score | 0.026728 (rank : 36) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P82198, Q3U9R1 | Gene names | Tgfbi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming growth factor-beta-induced protein ig-h3 precursor (Beta ig-h3). | |||||
|
IL4RA_MOUSE
|
||||||
| NC score | 0.026430 (rank : 37) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.016849 (rank : 38) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.013660 (rank : 39) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
CSPG5_MOUSE
|
||||||
| NC score | 0.010385 (rank : 40) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.010343 (rank : 41) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.009220 (rank : 42) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.004834 (rank : 43) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
MGR3_HUMAN
|
||||||
| NC score | 0.002990 (rank : 44) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14832, Q75MV4, Q75N17, Q86YG6, Q8TBH9 | Gene names | GRM3, GPRC1C, MGLUR3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | -0.001482 (rank : 45) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | -0.002358 (rank : 46) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||