Please be patient as the page loads
|
UGGG1_HUMAN
|
||||||
| SwissProt Accessions | Q9NYU2, Q8IW30, Q9H8I4 | Gene names | UGCGL1, GT, UGGT, UGT1, UGTR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 1 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase) (HUGT1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UGGG1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NYU2, Q8IW30, Q9H8I4 | Gene names | UGCGL1, GT, UGGT, UGT1, UGTR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 1 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase) (HUGT1). | |||||
|
UGGG1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997943 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P5E4, Q6NV70 | Gene names | Ugcgl1, Gt, Uggt, Ugt1, Ugtr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 1 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase). | |||||
|
UGGG2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.987845 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYU1, Q9UFC4 | Gene names | UGCGL2, UGT2 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 2 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase 2) (HUGT2). | |||||
|
DYN3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 4) | NC score | 0.030184 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
TOP2A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 5) | NC score | 0.028434 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11388, Q71UN1, Q9HB24, Q9HB25, Q9HB26, Q9UP44, Q9UQP9 | Gene names | TOP2A, TOP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
GGT1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 6) | NC score | 0.015309 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19440, Q08247, Q14404, Q8TBS1, Q9UMK1 | Gene names | GGT1, GGT | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-glutamyltranspeptidase 1 precursor (EC 2.3.2.2) (Gamma- glutamyltransferase 1) (GGT 1) (CD224 antigen) [Contains: Gamma- glutamyltranspeptidase 1 heavy chain; Gamma-glutamyltranspeptidase 1 light chain]. | |||||
|
LDHA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 7) | NC score | 0.014189 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00338, Q53G53, Q6IBM7, Q6ZNV1 | Gene names | LDHA, PIG19 | |||
|
Domain Architecture |
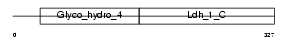
|
|||||
| Description | L-lactate dehydrogenase A chain (EC 1.1.1.27) (LDH-A) (LDH muscle subunit) (LDH-M) (Proliferation-inducing gene 19 protein) (NY-REN-59 antigen). | |||||
|
SPC24_HUMAN
|
||||||
| θ value | 3.0926 (rank : 8) | NC score | 0.019046 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NBT2 | Gene names | SPBC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc24 (hSpc24). | |||||
|
LARGE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 9) | NC score | 0.027533 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95461, O60348, Q9UGD1, Q9UGE7, Q9UGG3, Q9UGZ8, Q9UH22 | Gene names | LARGE, KIAA0609, LARGE1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycosyltransferase-like protein LARGE1 (EC 2.4.-.-) (Acetylglucosaminyltransferase-like 1A). | |||||
|
LARGE_MOUSE
|
||||||
| θ value | 4.03905 (rank : 10) | NC score | 0.028444 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1M7, Q497S9, Q6P7U2, Q80TW0 | Gene names | Large, Kiaa0609, Large1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycosyltransferase-like protein LARGE1 (EC 2.4.-.-) (Acetylglucosaminyltransferase-like 1A). | |||||
|
PRI2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 11) | NC score | 0.020117 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49643, Q53FJ8, Q6P1Q7, Q8WVL2, Q9H413 | Gene names | PRIM2A, PRIM2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA primase large subunit (EC 2.7.7.-) (DNA primase 58 kDa subunit) (p58). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 5.27518 (rank : 12) | NC score | 0.017322 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
PDP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 13) | NC score | 0.023815 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2J9 | Gene names | PDP2, KIAA1348 | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 2, mitochondrial precursor (EC 3.1.3.43) (PDP 2) (Pyruvate dehydrogenase phosphatase, catalytic subunit 2) (PDPC 2). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 14) | NC score | -0.000420 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
TTC7B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 15) | NC score | 0.012013 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86TV6, Q86U24, Q86VT3 | Gene names | TTC7B, TTC7L1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7B (Tetratricopeptide repeat protein 7-like-1). | |||||
|
CLSPN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 16) | NC score | 0.009651 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
I17RB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.021806 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JIP3, Q9JIP2 | Gene names | Il17rb, Evi27, Il17br | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-17 receptor B precursor (IL-17 receptor B) (IL-17RB) (Interleukin-17B receptor) (IL-17B receptor) (IL-17 receptor homolog 1) (IL-17Rh1) (IL17Rh1) (IL-17ER). | |||||
|
UGGG1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NYU2, Q8IW30, Q9H8I4 | Gene names | UGCGL1, GT, UGGT, UGT1, UGTR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 1 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase) (HUGT1). | |||||
|
UGGG1_MOUSE
|
||||||
| NC score | 0.997943 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P5E4, Q6NV70 | Gene names | Ugcgl1, Gt, Uggt, Ugt1, Ugtr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 1 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase). | |||||
|
UGGG2_HUMAN
|
||||||
| NC score | 0.987845 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYU1, Q9UFC4 | Gene names | UGCGL2, UGT2 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucose:glycoprotein glucosyltransferase 2 precursor (EC 2.4.1.-) (UDP-glucose ceramide glucosyltransferase-like 1) (UDP-- Glc:glycoprotein glucosyltransferase 2) (HUGT2). | |||||
|
DYN3_HUMAN
|
||||||
| NC score | 0.030184 (rank : 4) | θ value | 0.62314 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
LARGE_MOUSE
|
||||||
| NC score | 0.028444 (rank : 5) | θ value | 4.03905 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1M7, Q497S9, Q6P7U2, Q80TW0 | Gene names | Large, Kiaa0609, Large1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycosyltransferase-like protein LARGE1 (EC 2.4.-.-) (Acetylglucosaminyltransferase-like 1A). | |||||
|
TOP2A_HUMAN
|
||||||
| NC score | 0.028434 (rank : 6) | θ value | 1.06291 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11388, Q71UN1, Q9HB24, Q9HB25, Q9HB26, Q9UP44, Q9UQP9 | Gene names | TOP2A, TOP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
LARGE_HUMAN
|
||||||
| NC score | 0.027533 (rank : 7) | θ value | 4.03905 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95461, O60348, Q9UGD1, Q9UGE7, Q9UGG3, Q9UGZ8, Q9UH22 | Gene names | LARGE, KIAA0609, LARGE1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycosyltransferase-like protein LARGE1 (EC 2.4.-.-) (Acetylglucosaminyltransferase-like 1A). | |||||
|
PDP2_HUMAN
|
||||||
| NC score | 0.023815 (rank : 8) | θ value | 6.88961 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2J9 | Gene names | PDP2, KIAA1348 | |||
|
Domain Architecture |

|
|||||
| Description | [Pyruvate dehydrogenase [lipoamide]]-phosphatase 2, mitochondrial precursor (EC 3.1.3.43) (PDP 2) (Pyruvate dehydrogenase phosphatase, catalytic subunit 2) (PDPC 2). | |||||
|
I17RB_MOUSE
|
||||||
| NC score | 0.021806 (rank : 9) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JIP3, Q9JIP2 | Gene names | Il17rb, Evi27, Il17br | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-17 receptor B precursor (IL-17 receptor B) (IL-17RB) (Interleukin-17B receptor) (IL-17B receptor) (IL-17 receptor homolog 1) (IL-17Rh1) (IL17Rh1) (IL-17ER). | |||||
|
PRI2_HUMAN
|
||||||
| NC score | 0.020117 (rank : 10) | θ value | 4.03905 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49643, Q53FJ8, Q6P1Q7, Q8WVL2, Q9H413 | Gene names | PRIM2A, PRIM2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA primase large subunit (EC 2.7.7.-) (DNA primase 58 kDa subunit) (p58). | |||||
|
SPC24_HUMAN
|
||||||
| NC score | 0.019046 (rank : 11) | θ value | 3.0926 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NBT2 | Gene names | SPBC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc24 (hSpc24). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.017322 (rank : 12) | θ value | 5.27518 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
GGT1_HUMAN
|
||||||
| NC score | 0.015309 (rank : 13) | θ value | 2.36792 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19440, Q08247, Q14404, Q8TBS1, Q9UMK1 | Gene names | GGT1, GGT | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-glutamyltranspeptidase 1 precursor (EC 2.3.2.2) (Gamma- glutamyltransferase 1) (GGT 1) (CD224 antigen) [Contains: Gamma- glutamyltranspeptidase 1 heavy chain; Gamma-glutamyltranspeptidase 1 light chain]. | |||||
|
LDHA_HUMAN
|
||||||
| NC score | 0.014189 (rank : 14) | θ value | 2.36792 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00338, Q53G53, Q6IBM7, Q6ZNV1 | Gene names | LDHA, PIG19 | |||
|
Domain Architecture |

|
|||||
| Description | L-lactate dehydrogenase A chain (EC 1.1.1.27) (LDH-A) (LDH muscle subunit) (LDH-M) (Proliferation-inducing gene 19 protein) (NY-REN-59 antigen). | |||||
|
TTC7B_HUMAN
|
||||||
| NC score | 0.012013 (rank : 15) | θ value | 6.88961 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86TV6, Q86U24, Q86VT3 | Gene names | TTC7B, TTC7L1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 7B (Tetratricopeptide repeat protein 7-like-1). | |||||
|
CLSPN_MOUSE
|
||||||
| NC score | 0.009651 (rank : 16) | θ value | 8.99809 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | -0.000420 (rank : 17) | θ value | 6.88961 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||