Please be patient as the page loads
|
TPP2_HUMAN
|
||||||
| SwissProt Accessions | P29144 | Gene names | TPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tripeptidyl-peptidase 2 (EC 3.4.14.10) (Tripeptidyl-peptidase II) (TPP-II) (Tripeptidyl aminopeptidase). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TPP2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P29144 | Gene names | TPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tripeptidyl-peptidase 2 (EC 3.4.14.10) (Tripeptidyl-peptidase II) (TPP-II) (Tripeptidyl aminopeptidase). | |||||
|
TPP2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997785 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q64514 | Gene names | Tpp2 | |||
|
Domain Architecture |

|
|||||
| Description | Tripeptidyl-peptidase 2 (EC 3.4.14.10) (Tripeptidyl-peptidase II) (TPP-II) (Tripeptidyl aminopeptidase). | |||||
|
MBTP1_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 3) | NC score | 0.378130 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTZ2, Q6PG67 | Gene names | Mbtps1, S1p, Ski1 | |||
|
Domain Architecture |
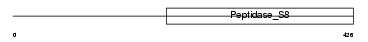
|
|||||
| Description | Membrane-bound transcription factor site 1 protease precursor (EC 3.4.21.-) (S1P endopeptidase) (Site-1 protease) (Subtilisin/kexin isozyme 1) (SKI-1) (Sterol-regulated luminal protease). | |||||
|
MBTP1_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 4) | NC score | 0.375467 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14703, Q9UF67 | Gene names | MBTPS1, KIAA0091, S1P, SKI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-bound transcription factor site 1 protease precursor (EC 3.4.21.-) (S1P endopeptidase) (Site-1 protease) (Subtilisin/kexin- isozyme 1) (SKI-1). | |||||
|
CSPG3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 5) | NC score | 0.020690 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
CNTN1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 6) | NC score | 0.012059 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P12960, Q6NXV7, Q8BR42, Q8C6A0 | Gene names | Cntn1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-1 precursor (Neural cell surface protein F3). | |||||
|
HECD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 7) | NC score | 0.026790 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
PTPRR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 8) | NC score | 0.015593 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15256, O00342, Q92682, Q9UE65 | Gene names | PTPRR, ECPTP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase PCPTP1) (NC-PTPCOM1) (Ch-1PTPase). | |||||
|
CNTN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 9) | NC score | 0.011198 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12860, Q12861, Q14030, Q8N466 | Gene names | CNTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-1 precursor (Neural cell surface protein F3) (Glycoprotein gp135). | |||||
|
PCSK9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 10) | NC score | 0.049684 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80W65, Q3UEH7, Q8BXW9, Q8CFT6 | Gene names | Pcsk9, Narc1 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 9 precursor (EC 3.4.21.-) (Proprotein convertase PC9) (Subtilisin/kexin-like protease PC9) (Neural apoptosis-regulated convertase 1) (NARC-1). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 11) | NC score | 0.006422 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.014435 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
GOGA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.009987 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
HCFC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.024210 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
HCFC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.023795 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.026139 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
PCSK9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.043048 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NBP7, Q5PSM5 | Gene names | PCSK9, NARC1 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 9 precursor (EC 3.4.21.-) (Proprotein convertase PC9) (Subtilisin/kexin-like protease PC9) (Neural apoptosis-regulated convertase 1) (NARC-1). | |||||
|
PP25_HUMAN
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.038348 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y547, Q9Y684 | Gene names | C1orf41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Placental protein 25 (PP25). | |||||
|
CRSP3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.024855 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULK4, O95403, Q9H0J2, Q9NTT9, Q9NTU0, Q9Y5P7, Q9Y667 | Gene names | CRSP3, ARC130, DRIP130, KIAA1216, SUR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 3 (Cofactor required for Sp1 transcriptional activation subunit 3) (Transcriptional coactivator CRSP130) (Vitamin D3 receptor-interacting protein complex 130 kDa component) (DRIP130) (Activator-recruited cofactor 130 kDa component) (ARC130). | |||||
|
MYG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.021505 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HB07 | Gene names | C12orf10 | |||
|
Domain Architecture |
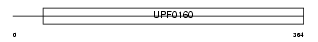
|
|||||
| Description | UPF0160 protein MYG1. | |||||
|
PTN13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.009607 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
LPPRC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.031446 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42704 | Gene names | LRPPRC, LRP130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 130 kDa leucine-rich protein (LRP 130) (GP130) (Leucine-rich PPR motif-containing protein). | |||||
|
MORC4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.011627 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TE76, Q5JUK7, Q96MZ2, Q9HAI7 | Gene names | MORC4, ZCWCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
ORC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.017172 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60862 | Gene names | Orc2l, Orc2 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 2. | |||||
|
PP25_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.032752 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D6H2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Placental protein 25 homolog (PP25). | |||||
|
AL3B2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.015708 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48448, Q8NAL5, Q96IB2 | Gene names | ALDH3B2, ALDH8 | |||
|
Domain Architecture |
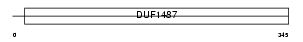
|
|||||
| Description | Aldehyde dehydrogenase 3B2 (EC 1.2.1.5) (Aldehyde dehydrogenase 8). | |||||
|
CK5P2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.008452 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.010962 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
LPXN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.009865 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99N69 | Gene names | Lpxn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leupaxin. | |||||
|
SULF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.008500 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWU6, Q86YV8, Q8NCA2, Q9UPS5 | Gene names | SULF1, KIAA1077 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular sulfatase Sulf-1 precursor (EC 3.1.6.-) (HSulf-1). | |||||
|
TPP2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P29144 | Gene names | TPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tripeptidyl-peptidase 2 (EC 3.4.14.10) (Tripeptidyl-peptidase II) (TPP-II) (Tripeptidyl aminopeptidase). | |||||
|
TPP2_MOUSE
|
||||||
| NC score | 0.997785 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q64514 | Gene names | Tpp2 | |||
|
Domain Architecture |

|
|||||
| Description | Tripeptidyl-peptidase 2 (EC 3.4.14.10) (Tripeptidyl-peptidase II) (TPP-II) (Tripeptidyl aminopeptidase). | |||||
|
MBTP1_MOUSE
|
||||||
| NC score | 0.378130 (rank : 3) | θ value | 3.89403e-11 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTZ2, Q6PG67 | Gene names | Mbtps1, S1p, Ski1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-bound transcription factor site 1 protease precursor (EC 3.4.21.-) (S1P endopeptidase) (Site-1 protease) (Subtilisin/kexin isozyme 1) (SKI-1) (Sterol-regulated luminal protease). | |||||
|
MBTP1_HUMAN
|
||||||
| NC score | 0.375467 (rank : 4) | θ value | 6.64225e-11 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14703, Q9UF67 | Gene names | MBTPS1, KIAA0091, S1P, SKI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-bound transcription factor site 1 protease precursor (EC 3.4.21.-) (S1P endopeptidase) (Site-1 protease) (Subtilisin/kexin- isozyme 1) (SKI-1). | |||||
|
PCSK9_MOUSE
|
||||||
| NC score | 0.049684 (rank : 5) | θ value | 2.36792 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80W65, Q3UEH7, Q8BXW9, Q8CFT6 | Gene names | Pcsk9, Narc1 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 9 precursor (EC 3.4.21.-) (Proprotein convertase PC9) (Subtilisin/kexin-like protease PC9) (Neural apoptosis-regulated convertase 1) (NARC-1). | |||||
|
PCSK9_HUMAN
|
||||||
| NC score | 0.043048 (rank : 6) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NBP7, Q5PSM5 | Gene names | PCSK9, NARC1 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 9 precursor (EC 3.4.21.-) (Proprotein convertase PC9) (Subtilisin/kexin-like protease PC9) (Neural apoptosis-regulated convertase 1) (NARC-1). | |||||
|
PP25_HUMAN
|
||||||
| NC score | 0.038348 (rank : 7) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y547, Q9Y684 | Gene names | C1orf41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Placental protein 25 (PP25). | |||||
|
PP25_MOUSE
|
||||||
| NC score | 0.032752 (rank : 8) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D6H2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Placental protein 25 homolog (PP25). | |||||
|
LPPRC_HUMAN
|
||||||
| NC score | 0.031446 (rank : 9) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42704 | Gene names | LRPPRC, LRP130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 130 kDa leucine-rich protein (LRP 130) (GP130) (Leucine-rich PPR motif-containing protein). | |||||
|
HECD1_HUMAN
|
||||||
| NC score | 0.026790 (rank : 10) | θ value | 1.38821 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.026139 (rank : 11) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
CRSP3_HUMAN
|
||||||
| NC score | 0.024855 (rank : 12) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULK4, O95403, Q9H0J2, Q9NTT9, Q9NTU0, Q9Y5P7, Q9Y667 | Gene names | CRSP3, ARC130, DRIP130, KIAA1216, SUR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 3 (Cofactor required for Sp1 transcriptional activation subunit 3) (Transcriptional coactivator CRSP130) (Vitamin D3 receptor-interacting protein complex 130 kDa component) (DRIP130) (Activator-recruited cofactor 130 kDa component) (ARC130). | |||||
|
HCFC1_HUMAN
|
||||||
| NC score | 0.024210 (rank : 13) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
HCFC1_MOUSE
|
||||||
| NC score | 0.023795 (rank : 14) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
MYG1_HUMAN
|
||||||
| NC score | 0.021505 (rank : 15) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HB07 | Gene names | C12orf10 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0160 protein MYG1. | |||||
|
CSPG3_MOUSE
|
||||||
| NC score | 0.020690 (rank : 16) | θ value | 0.813845 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
ORC2_MOUSE
|
||||||
| NC score | 0.017172 (rank : 17) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60862 | Gene names | Orc2l, Orc2 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 2. | |||||
|
AL3B2_HUMAN
|
||||||
| NC score | 0.015708 (rank : 18) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48448, Q8NAL5, Q96IB2 | Gene names | ALDH3B2, ALDH8 | |||
|
Domain Architecture |

|
|||||
| Description | Aldehyde dehydrogenase 3B2 (EC 1.2.1.5) (Aldehyde dehydrogenase 8). | |||||
|
PTPRR_HUMAN
|
||||||
| NC score | 0.015593 (rank : 19) | θ value | 1.81305 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15256, O00342, Q92682, Q9UE65 | Gene names | PTPRR, ECPTP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase PCPTP1) (NC-PTPCOM1) (Ch-1PTPase). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.014435 (rank : 20) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CNTN1_MOUSE
|
||||||
| NC score | 0.012059 (rank : 21) | θ value | 1.38821 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P12960, Q6NXV7, Q8BR42, Q8C6A0 | Gene names | Cntn1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-1 precursor (Neural cell surface protein F3). | |||||
|
MORC4_HUMAN
|
||||||
| NC score | 0.011627 (rank : 22) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TE76, Q5JUK7, Q96MZ2, Q9HAI7 | Gene names | MORC4, ZCWCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 4 (Zinc finger CW-type coiled- coil domain protein 2). | |||||
|
CNTN1_HUMAN
|
||||||
| NC score | 0.011198 (rank : 23) | θ value | 2.36792 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12860, Q12861, Q14030, Q8N466 | Gene names | CNTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-1 precursor (Neural cell surface protein F3) (Glycoprotein gp135). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.010962 (rank : 24) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
GOGA2_HUMAN
|
||||||
| NC score | 0.009987 (rank : 25) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
LPXN_MOUSE
|
||||||
| NC score | 0.009865 (rank : 26) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99N69 | Gene names | Lpxn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leupaxin. | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.009607 (rank : 27) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
SULF1_HUMAN
|
||||||
| NC score | 0.008500 (rank : 28) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWU6, Q86YV8, Q8NCA2, Q9UPS5 | Gene names | SULF1, KIAA1077 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular sulfatase Sulf-1 precursor (EC 3.1.6.-) (HSulf-1). | |||||
|
CK5P2_MOUSE
|
||||||
| NC score | 0.008452 (rank : 29) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.006422 (rank : 30) | θ value | 3.0926 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||