Please be patient as the page loads
|
TM55B_MOUSE
|
||||||
| SwissProt Accessions | Q3TWL2 | Gene names | Tmem55b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 55B (EC 3.1.3.-) (Type I phosphatidylinositol 4,5-bisphosphate 4-phosphatase) (PtdIns-4,5-P2 4-Ptase I). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TM55B_MOUSE
|
||||||
| θ value | 1.05497e-141 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q3TWL2 | Gene names | Tmem55b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 55B (EC 3.1.3.-) (Type I phosphatidylinositol 4,5-bisphosphate 4-phosphatase) (PtdIns-4,5-P2 4-Ptase I). | |||||
|
TM55B_HUMAN
|
||||||
| θ value | 1.16911e-132 (rank : 2) | NC score | 0.986908 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86T03, Q86U09, Q8WUC0, Q9BU67, Q9NSU8 | Gene names | TMEM55B, C14orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 55B (EC 3.1.3.-) (Type I phosphatidylinositol 4,5-bisphosphate 4-phosphatase) (PtdIns-4,5-P2 4-Ptase I). | |||||
|
TM55A_HUMAN
|
||||||
| θ value | 1.18825e-76 (rank : 3) | NC score | 0.961649 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N4L2, Q68CU2 | Gene names | TMEM55A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 55A (EC 3.1.3.-) (Type II phosphatidylinositol 4,5-bisphosphate 4-phosphatase) (PtdIns-4,5-P2 4-Ptase II). | |||||
|
TM55A_MOUSE
|
||||||
| θ value | 2.92676e-75 (rank : 4) | NC score | 0.960500 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CZX7 | Gene names | Tmem55a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 55A (EC 3.1.3.-) (Type II phosphatidylinositol 4,5-bisphosphate 4-phosphatase) (PtdIns-4,5-P2 4-Ptase II). | |||||
|
SOX15_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 5) | NC score | 0.033979 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43267, O70204, P70418, Q62246, Q91V00, Q91V43, Q920T1, Q9JLG2 | Gene names | Sox15, Sox-15 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein. | |||||
|
EGR4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 6) | NC score | 0.015455 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
FRAS1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 7) | NC score | 0.047959 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
FBLI1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 8) | NC score | 0.026314 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q71FD7, Q99J35 | Gene names | Fblim1, Cal | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-binding LIM protein 1 (CSX-associated LIM). | |||||
|
HNRL1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 9) | NC score | 0.050967 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
TMM28_HUMAN
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.074307 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75949 | Gene names | TMEM28, TED | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 28 (TED protein). | |||||
|
JPH2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.035794 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
NMDE3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.018923 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.050235 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
IL4RA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 14) | NC score | 0.037906 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P24394, Q96P01, Q9H181, Q9H182, Q9H183, Q9H184, Q9H185, Q9H186, Q9H187, Q9H188 | Gene names | IL4R, 582J2.1, IL4RA | |||
|
Domain Architecture |
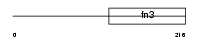
|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (sIL4Ralpha/prot) (IL-4-binding protein) (IL4-BP)]. | |||||
|
MAGC3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.021454 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TD91, Q5JZ43, Q9BZ80 | Gene names | MAGEC3, HCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C3 (MAGE-C3 antigen) (Hepatocellular carcinoma-associated antigen 2). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.026856 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
PER1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.020528 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.013667 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.046998 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.023728 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
HNRL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.042954 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BUJ2, O76022, Q6ZSZ0, Q7L8P4, Q8N6Z4, Q96G37, Q9HAL3, Q9UG75 | Gene names | HNRPUL1, E1BAP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1 (Adenovirus early region 1B-associated protein 5) (E1B-55 kDa-associated protein 5) (E1B-AP5). | |||||
|
FMNL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.017627 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.016727 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
ZN628_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.001470 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5EBL2, Q86X34 | Gene names | ZNF628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628. | |||||
|
ADA29_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.005801 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKF5, Q9UHP1, Q9UKF3, Q9UKF4 | Gene names | ADAM29 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 29 precursor (A disintegrin and metalloproteinase domain 29). | |||||
|
CD2L7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.001090 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
HXA3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.005550 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P02831, Q4V9Z8, Q61197 | Gene names | Hoxa3, Hox-1.5, Hoxa-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1.5) (MO-10). | |||||
|
PRP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.020282 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.020727 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RIN3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.016081 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TB24, Q8NF30, Q8TEE8, Q8WYP4, Q9H6A5, Q9HAG1 | Gene names | RIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
SMR3B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.045552 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P02814, Q9UBN0, Q9UCT0 | Gene names | SMR3B, PBII, PROL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog B precursor (Proline-rich protein 3) (Proline-rich peptide P-B) [Contains: Peptide P-A; Peptide D1A]. | |||||
|
SNIP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.017439 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9C0H9, Q8N4W8 | Gene names | P140, KIAA1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
SREC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.010992 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.011699 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
ENAM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.023351 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
IQEC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.015533 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.016316 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IRX5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.010379 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78411, P78416, Q7Z2E1 | Gene names | IRX5, IRX2A, IRXB2 | |||
|
Domain Architecture |
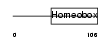
|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2) (IRX-2A). | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.019550 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.002677 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
SMR3A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.032656 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
WASP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.015901 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
F8I2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.015352 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23610, Q8IXP3 | Gene names | F8A1, F8A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor VIII intron 22 protein (CpG island protein). | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.005331 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
FRAS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.026318 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.015960 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MILK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.008698 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.009464 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SNX18_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.008462 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RF0 | Gene names | SNAG1, SNX18 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-18 (Sorting nexin-associated Golgi protein 1). | |||||
|
ZBT45_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.002570 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96K62 | Gene names | ZBTB45, ZNF499 | |||
|
Domain Architecture |
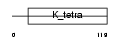
|
|||||
| Description | Zinc finger and BTB domain-containing protein 45 (Zinc finger protein 499). | |||||
|
TM55B_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.05497e-141 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q3TWL2 | Gene names | Tmem55b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 55B (EC 3.1.3.-) (Type I phosphatidylinositol 4,5-bisphosphate 4-phosphatase) (PtdIns-4,5-P2 4-Ptase I). | |||||
|
TM55B_HUMAN
|
||||||
| NC score | 0.986908 (rank : 2) | θ value | 1.16911e-132 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86T03, Q86U09, Q8WUC0, Q9BU67, Q9NSU8 | Gene names | TMEM55B, C14orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 55B (EC 3.1.3.-) (Type I phosphatidylinositol 4,5-bisphosphate 4-phosphatase) (PtdIns-4,5-P2 4-Ptase I). | |||||
|
TM55A_HUMAN
|
||||||
| NC score | 0.961649 (rank : 3) | θ value | 1.18825e-76 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N4L2, Q68CU2 | Gene names | TMEM55A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 55A (EC 3.1.3.-) (Type II phosphatidylinositol 4,5-bisphosphate 4-phosphatase) (PtdIns-4,5-P2 4-Ptase II). | |||||
|
TM55A_MOUSE
|
||||||
| NC score | 0.960500 (rank : 4) | θ value | 2.92676e-75 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CZX7 | Gene names | Tmem55a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 55A (EC 3.1.3.-) (Type II phosphatidylinositol 4,5-bisphosphate 4-phosphatase) (PtdIns-4,5-P2 4-Ptase II). | |||||
|
TMM28_HUMAN
|
||||||
| NC score | 0.074307 (rank : 5) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75949 | Gene names | TMEM28, TED | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 28 (TED protein). | |||||
|
HNRL1_MOUSE
|
||||||
| NC score | 0.050967 (rank : 6) | θ value | 0.279714 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.050235 (rank : 7) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
FRAS1_MOUSE
|
||||||
| NC score | 0.047959 (rank : 8) | θ value | 0.0736092 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.046998 (rank : 9) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
SMR3B_HUMAN
|
||||||
| NC score | 0.045552 (rank : 10) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P02814, Q9UBN0, Q9UCT0 | Gene names | SMR3B, PBII, PROL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog B precursor (Proline-rich protein 3) (Proline-rich peptide P-B) [Contains: Peptide P-A; Peptide D1A]. | |||||
|
HNRL1_HUMAN
|
||||||
| NC score | 0.042954 (rank : 11) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BUJ2, O76022, Q6ZSZ0, Q7L8P4, Q8N6Z4, Q96G37, Q9HAL3, Q9UG75 | Gene names | HNRPUL1, E1BAP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1 (Adenovirus early region 1B-associated protein 5) (E1B-55 kDa-associated protein 5) (E1B-AP5). | |||||
|
IL4RA_HUMAN
|
||||||
| NC score | 0.037906 (rank : 12) | θ value | 1.06291 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P24394, Q96P01, Q9H181, Q9H182, Q9H183, Q9H184, Q9H185, Q9H186, Q9H187, Q9H188 | Gene names | IL4R, 582J2.1, IL4RA | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (sIL4Ralpha/prot) (IL-4-binding protein) (IL4-BP)]. | |||||
|
JPH2_MOUSE
|
||||||
| NC score | 0.035794 (rank : 13) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ET78, Q9ET79 | Gene names | Jph2, Jp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
SOX15_MOUSE
|
||||||
| NC score | 0.033979 (rank : 14) | θ value | 0.0563607 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43267, O70204, P70418, Q62246, Q91V00, Q91V43, Q920T1, Q9JLG2 | Gene names | Sox15, Sox-15 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-15 protein. | |||||
|
SMR3A_HUMAN
|
||||||
| NC score | 0.032656 (rank : 15) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.026856 (rank : 16) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
FRAS1_HUMAN
|
||||||
| NC score | 0.026318 (rank : 17) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
FBLI1_MOUSE
|
||||||
| NC score | 0.026314 (rank : 18) | θ value | 0.21417 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q71FD7, Q99J35 | Gene names | Fblim1, Cal | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-binding LIM protein 1 (CSX-associated LIM). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.023728 (rank : 19) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
ENAM_HUMAN
|
||||||
| NC score | 0.023351 (rank : 20) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
MAGC3_HUMAN
|
||||||
| NC score | 0.021454 (rank : 21) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TD91, Q5JZ43, Q9BZ80 | Gene names | MAGEC3, HCA2 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C3 (MAGE-C3 antigen) (Hepatocellular carcinoma-associated antigen 2). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.020727 (rank : 22) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.020528 (rank : 23) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.020282 (rank : 24) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.019550 (rank : 25) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
NMDE3_HUMAN
|
||||||
| NC score | 0.018923 (rank : 26) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14957 | Gene names | GRIN2C | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate [NMDA] receptor subunit epsilon 3 precursor (N-methyl D- aspartate receptor subtype 2C) (NR2C) (NMDAR2C). | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.017627 (rank : 27) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
SNIP_HUMAN
|
||||||
| NC score | 0.017439 (rank : 28) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9C0H9, Q8N4W8 | Gene names | P140, KIAA1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.016727 (rank : 29) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
IQEC2_MOUSE
|
||||||
| NC score | 0.016316 (rank : 30) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
RIN3_HUMAN
|
||||||
| NC score | 0.016081 (rank : 31) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TB24, Q8NF30, Q8TEE8, Q8WYP4, Q9H6A5, Q9HAG1 | Gene names | RIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.015960 (rank : 32) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
WASP_MOUSE
|
||||||
| NC score | 0.015901 (rank : 33) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
IQEC2_HUMAN
|
||||||
| NC score | 0.015533 (rank : 34) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
EGR4_HUMAN
|
||||||
| NC score | 0.015455 (rank : 35) | θ value | 0.0736092 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
F8I2_HUMAN
|
||||||
| NC score | 0.015352 (rank : 36) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23610, Q8IXP3 | Gene names | F8A1, F8A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor VIII intron 22 protein (CpG island protein). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.013667 (rank : 37) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.011699 (rank : 38) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.010992 (rank : 39) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
IRX5_HUMAN
|
||||||
| NC score | 0.010379 (rank : 40) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78411, P78416, Q7Z2E1 | Gene names | IRX5, IRX2A, IRXB2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2) (IRX-2A). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.009464 (rank : 41) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MILK1_HUMAN
|
||||||
| NC score | 0.008698 (rank : 42) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
SNX18_HUMAN
|
||||||
| NC score | 0.008462 (rank : 43) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RF0 | Gene names | SNAG1, SNX18 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-18 (Sorting nexin-associated Golgi protein 1). | |||||
|
ADA29_HUMAN
|
||||||
| NC score | 0.005801 (rank : 44) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UKF5, Q9UHP1, Q9UKF3, Q9UKF4 | Gene names | ADAM29 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 29 precursor (A disintegrin and metalloproteinase domain 29). | |||||
|
HXA3_MOUSE
|
||||||
| NC score | 0.005550 (rank : 45) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P02831, Q4V9Z8, Q61197 | Gene names | Hoxa3, Hox-1.5, Hoxa-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1.5) (MO-10). | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.005331 (rank : 46) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.002677 (rank : 47) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
ZBT45_HUMAN
|
||||||
| NC score | 0.002570 (rank : 48) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96K62 | Gene names | ZBTB45, ZNF499 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 45 (Zinc finger protein 499). | |||||
|
ZN628_HUMAN
|
||||||
| NC score | 0.001470 (rank : 49) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5EBL2, Q86X34 | Gene names | ZNF628 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 628. | |||||
|
CD2L7_HUMAN
|
||||||
| NC score | 0.001090 (rank : 50) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||