Please be patient as the page loads
|
TGM5_HUMAN
|
||||||
| SwissProt Accessions | O43548, O43549, Q9UEZ4 | Gene names | TGM5, TGMX | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase 5 (EC 2.3.2.13) (Transglutaminase-5) (TGase 5) (Transglutaminase X) (TGase X) (TGX) (TG(X)). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TGM5_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43548, O43549, Q9UEZ4 | Gene names | TGM5, TGMX | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase 5 (EC 2.3.2.13) (Transglutaminase-5) (TGase 5) (Transglutaminase X) (TGase X) (TGX) (TG(X)). | |||||
|
TGM5_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998971 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D7I9, Q3V1F9 | Gene names | Tgm5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-glutamine gamma-glutamyltransferase 5 (EC 2.3.2.13) (Transglutaminase-5) (TGase 5). | |||||
|
TGM7_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.993407 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96PF1 | Gene names | TGM7 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase Z (EC 2.3.2.13) (TGase Z) (TGZ) (TG(Z)) (Transglutaminase 7). | |||||
|
TGM3L_HUMAN
|
||||||
| θ value | 1.40939e-178 (rank : 4) | NC score | 0.991925 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95932, Q5JXU4, Q5JXU5, Q719M2, Q719M3, Q9Y4U8 | Gene names | TGM6, TGM3L | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase 6 (EC 2.3.2.13) (Transglutaminase-3-like) (TGase-3-like) (Transglutaminase Y). | |||||
|
TGM3_HUMAN
|
||||||
| θ value | 3.97187e-173 (rank : 5) | NC score | 0.990204 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08188, O95933 | Gene names | TGM3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase E precursor (EC 2.3.2.13) (TGase E) (TGE) (TG(E)) (Transglutaminase-3) [Contains: Protein- glutamine gamma-glutamyltransferase E 50 kDa non-catalytic chain; Protein-glutamine gamma-glutamyltransferase E 27 kDa catalytic chain]. | |||||
|
TGM3_MOUSE
|
||||||
| θ value | 2.94559e-168 (rank : 6) | NC score | 0.989649 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08189 | Gene names | Tgm3, Tgase3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase E precursor (EC 2.3.2.13) (TGase E) (TGE) (TG(E)) (Transglutaminase-3) [Contains: Protein- glutamine gamma-glutamyltransferase E 50 kDa non-catalytic chain; Protein-glutamine gamma-glutamyltransferase E 27 kDa catalytic chain]. | |||||
|
TGM2_MOUSE
|
||||||
| θ value | 4.26331e-159 (rank : 7) | NC score | 0.987487 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P21981, O88901, Q9R1F7 | Gene names | Tgm2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase 2 (EC 2.3.2.13) (Tissue transglutaminase) (TGase C) (TGC) (TG(C)) (Transglutaminase-2). | |||||
|
TGM2_HUMAN
|
||||||
| θ value | 2.04937e-153 (rank : 8) | NC score | 0.986721 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P21980, Q16436, Q9BTN7, Q9UH35 | Gene names | TGM2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase 2 (EC 2.3.2.13) (Tissue transglutaminase) (TGase C) (TGC) (TG(C)) (Transglutaminase-2) (TGase- H). | |||||
|
TGM1_MOUSE
|
||||||
| θ value | 4.30305e-127 (rank : 9) | NC score | 0.975884 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JLF6, Q8R0T9 | Gene names | Tgm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-glutamine gamma-glutamyltransferase K (EC 2.3.2.13) (Transglutaminase K) (TGase K) (TGK) (TG(K)) (Transglutaminase-1) (Epidermal TGase). | |||||
|
TGM1_HUMAN
|
||||||
| θ value | 4.02752e-125 (rank : 10) | NC score | 0.975702 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P22735 | Gene names | TGM1, KTG | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase K (EC 2.3.2.13) (Transglutaminase K) (TGase K) (TGK) (TG(K)) (Transglutaminase-1) (Epidermal TGase). | |||||
|
F13A_MOUSE
|
||||||
| θ value | 2.89301e-115 (rank : 11) | NC score | 0.971278 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BH61, Q3TNF9, Q8BIP2 | Gene names | F13a1, F13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coagulation factor XIII A chain precursor (EC 2.3.2.13) (Coagulation factor XIIIa) (Protein-glutamine gamma-glutamyltransferase A chain) (Transglutaminase A chain). | |||||
|
F13A_HUMAN
|
||||||
| θ value | 1.02895e-112 (rank : 12) | NC score | 0.971339 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00488, Q8N6X2, Q96P24 | Gene names | F13A1, F13A | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII A chain precursor (EC 2.3.2.13) (Coagulation factor XIIIa) (Protein-glutamine gamma-glutamyltransferase A chain) (Transglutaminase A chain). | |||||
|
TGM4_HUMAN
|
||||||
| θ value | 2.89973e-107 (rank : 13) | NC score | 0.979797 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49221, Q16707, Q96QN4 | Gene names | TGM4 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase 4 (EC 2.3.2.13) (Transglutaminase-4) (TGase-4) (Prostate transglutaminase) (TGP) (TG(P)) (Prostate-specific transglutaminase) (Fibrinoligase). | |||||
|
EPB42_HUMAN
|
||||||
| θ value | 8.43692e-107 (rank : 14) | NC score | 0.971376 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P16452 | Gene names | EPB42, E42P | |||
|
Domain Architecture |

|
|||||
| Description | Erythrocyte membrane protein band 4.2 (Erythrocyte protein 4.2) (P4.2). | |||||
|
EPB42_MOUSE
|
||||||
| θ value | 1.34698e-104 (rank : 15) | NC score | 0.969926 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49222 | Gene names | Epb42, Epb4.2 | |||
|
Domain Architecture |

|
|||||
| Description | Erythrocyte membrane protein band 4.2 (Erythrocyte protein 4.2) (P4.2). | |||||
|
K1210_HUMAN
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.021205 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
HS74L_MOUSE
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.012333 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48722, P97854, Q8BQD0, Q8CC45, Q91X29 | Gene names | Hspa4l, Apg1, Hsp4l, Osp94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
NPM_MOUSE
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.015896 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61937 | Gene names | Npm1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38). | |||||
|
UB2D1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.007004 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51668 | Gene names | UBE2D1, UBCH5, UBCH5A | |||
|
Domain Architecture |
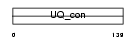
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 D1 (EC 6.3.2.19) (Ubiquitin-protein ligase D1) (Ubiquitin carrier protein D1) (UbcH5) (Ubiquitin- conjugating enzyme E2-17 kDa 1) (E2(17)KB 1). | |||||
|
UB2D1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.007004 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61080 | Gene names | Ube2d1 | |||
|
Domain Architecture |
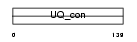
|
|||||
| Description | Ubiquitin-conjugating enzyme E2 D1 (EC 6.3.2.19) (Ubiquitin-protein ligase D1) (Ubiquitin carrier protein D1) (Ubiquitin-conjugating enzyme E2-17 kDa 1) (E2(17)KB 1). | |||||
|
2A5B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.007212 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15173, Q13853 | Gene names | PPP2R5B | |||
|
Domain Architecture |
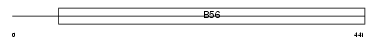
|
|||||
| Description | Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit beta isoform (PP2A, B subunit, B' beta isoform) (PP2A, B subunit, B56 beta isoform) (PP2A, B subunit, PR61 beta isoform) (PP2A, B subunit, R5 beta isoform). | |||||
|
CLSPN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.010712 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
HS74L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.008930 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95757, Q4W5M5, Q8IWA2 | Gene names | HSPA4L, APG1, OSP94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.006503 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
CHK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | -0.002508 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O96017, Q6QA03, Q6QA04, Q6QA05, Q6QA06, Q6QA07, Q6QA08, Q6QA10, Q6QA11, Q6QA12, Q6QA13, Q9HCQ8, Q9UGF0, Q9UGF1 | Gene names | CHEK2, CHK2 | |||
|
Domain Architecture |
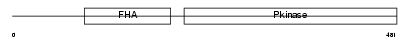
|
|||||
| Description | Serine/threonine-protein kinase Chk2 (EC 2.7.11.1) (Cds1). | |||||
|
QPCT_MOUSE
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.011213 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CYK2, Q8BH20, Q8VE07 | Gene names | Qpct | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutaminyl-peptide cyclotransferase precursor (EC 2.3.2.5) (QC) (Glutaminyl-tRNA cyclotransferase) (Glutaminyl cyclase). | |||||
|
SM1L2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.000340 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
HSP74_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.007091 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
LRRC4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | -0.000394 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBW1, Q6ZMI8, Q96A85 | Gene names | LRRC4, BAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 4 precursor (Brain tumor- associated protein LRRC4) (NAG14). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | -0.000662 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
TGM5_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43548, O43549, Q9UEZ4 | Gene names | TGM5, TGMX | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase 5 (EC 2.3.2.13) (Transglutaminase-5) (TGase 5) (Transglutaminase X) (TGase X) (TGX) (TG(X)). | |||||
|
TGM5_MOUSE
|
||||||
| NC score | 0.998971 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D7I9, Q3V1F9 | Gene names | Tgm5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-glutamine gamma-glutamyltransferase 5 (EC 2.3.2.13) (Transglutaminase-5) (TGase 5). | |||||
|
TGM7_HUMAN
|
||||||
| NC score | 0.993407 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96PF1 | Gene names | TGM7 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase Z (EC 2.3.2.13) (TGase Z) (TGZ) (TG(Z)) (Transglutaminase 7). | |||||
|
TGM3L_HUMAN
|
||||||
| NC score | 0.991925 (rank : 4) | θ value | 1.40939e-178 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95932, Q5JXU4, Q5JXU5, Q719M2, Q719M3, Q9Y4U8 | Gene names | TGM6, TGM3L | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase 6 (EC 2.3.2.13) (Transglutaminase-3-like) (TGase-3-like) (Transglutaminase Y). | |||||
|
TGM3_HUMAN
|
||||||
| NC score | 0.990204 (rank : 5) | θ value | 3.97187e-173 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08188, O95933 | Gene names | TGM3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase E precursor (EC 2.3.2.13) (TGase E) (TGE) (TG(E)) (Transglutaminase-3) [Contains: Protein- glutamine gamma-glutamyltransferase E 50 kDa non-catalytic chain; Protein-glutamine gamma-glutamyltransferase E 27 kDa catalytic chain]. | |||||
|
TGM3_MOUSE
|
||||||
| NC score | 0.989649 (rank : 6) | θ value | 2.94559e-168 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08189 | Gene names | Tgm3, Tgase3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase E precursor (EC 2.3.2.13) (TGase E) (TGE) (TG(E)) (Transglutaminase-3) [Contains: Protein- glutamine gamma-glutamyltransferase E 50 kDa non-catalytic chain; Protein-glutamine gamma-glutamyltransferase E 27 kDa catalytic chain]. | |||||
|
TGM2_MOUSE
|
||||||
| NC score | 0.987487 (rank : 7) | θ value | 4.26331e-159 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P21981, O88901, Q9R1F7 | Gene names | Tgm2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase 2 (EC 2.3.2.13) (Tissue transglutaminase) (TGase C) (TGC) (TG(C)) (Transglutaminase-2). | |||||
|
TGM2_HUMAN
|
||||||
| NC score | 0.986721 (rank : 8) | θ value | 2.04937e-153 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P21980, Q16436, Q9BTN7, Q9UH35 | Gene names | TGM2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase 2 (EC 2.3.2.13) (Tissue transglutaminase) (TGase C) (TGC) (TG(C)) (Transglutaminase-2) (TGase- H). | |||||
|
TGM4_HUMAN
|
||||||
| NC score | 0.979797 (rank : 9) | θ value | 2.89973e-107 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49221, Q16707, Q96QN4 | Gene names | TGM4 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase 4 (EC 2.3.2.13) (Transglutaminase-4) (TGase-4) (Prostate transglutaminase) (TGP) (TG(P)) (Prostate-specific transglutaminase) (Fibrinoligase). | |||||
|
TGM1_MOUSE
|
||||||
| NC score | 0.975884 (rank : 10) | θ value | 4.30305e-127 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JLF6, Q8R0T9 | Gene names | Tgm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-glutamine gamma-glutamyltransferase K (EC 2.3.2.13) (Transglutaminase K) (TGase K) (TGK) (TG(K)) (Transglutaminase-1) (Epidermal TGase). | |||||
|
TGM1_HUMAN
|
||||||
| NC score | 0.975702 (rank : 11) | θ value | 4.02752e-125 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P22735 | Gene names | TGM1, KTG | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase K (EC 2.3.2.13) (Transglutaminase K) (TGase K) (TGK) (TG(K)) (Transglutaminase-1) (Epidermal TGase). | |||||
|
EPB42_HUMAN
|
||||||
| NC score | 0.971376 (rank : 12) | θ value | 8.43692e-107 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P16452 | Gene names | EPB42, E42P | |||
|
Domain Architecture |

|
|||||
| Description | Erythrocyte membrane protein band 4.2 (Erythrocyte protein 4.2) (P4.2). | |||||
|
F13A_HUMAN
|
||||||
| NC score | 0.971339 (rank : 13) | θ value | 1.02895e-112 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00488, Q8N6X2, Q96P24 | Gene names | F13A1, F13A | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XIII A chain precursor (EC 2.3.2.13) (Coagulation factor XIIIa) (Protein-glutamine gamma-glutamyltransferase A chain) (Transglutaminase A chain). | |||||
|
F13A_MOUSE
|
||||||
| NC score | 0.971278 (rank : 14) | θ value | 2.89301e-115 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BH61, Q3TNF9, Q8BIP2 | Gene names | F13a1, F13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coagulation factor XIII A chain precursor (EC 2.3.2.13) (Coagulation factor XIIIa) (Protein-glutamine gamma-glutamyltransferase A chain) (Transglutaminase A chain). | |||||
|
EPB42_MOUSE
|
||||||
| NC score | 0.969926 (rank : 15) | θ value | 1.34698e-104 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P49222 | Gene names | Epb42, Epb4.2 | |||
|
Domain Architecture |

|
|||||
| Description | Erythrocyte membrane protein band 4.2 (Erythrocyte protein 4.2) (P4.2). | |||||
|
K1210_HUMAN
|
||||||
| NC score | 0.021205 (rank : 16) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
NPM_MOUSE
|
||||||
| NC score | 0.015896 (rank : 17) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61937 | Gene names | Npm1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38). | |||||
|
HS74L_MOUSE
|
||||||
| NC score | 0.012333 (rank : 18) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48722, P97854, Q8BQD0, Q8CC45, Q91X29 | Gene names | Hspa4l, Apg1, Hsp4l, Osp94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
QPCT_MOUSE
|
||||||
| NC score | 0.011213 (rank : 19) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CYK2, Q8BH20, Q8VE07 | Gene names | Qpct | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutaminyl-peptide cyclotransferase precursor (EC 2.3.2.5) (QC) (Glutaminyl-tRNA cyclotransferase) (Glutaminyl cyclase). | |||||
|
CLSPN_MOUSE
|
||||||
| NC score | 0.010712 (rank : 20) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80YR7, Q69GM2 | Gene names | Clspn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin. | |||||
|
HS74L_HUMAN
|
||||||
| NC score | 0.008930 (rank : 21) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95757, Q4W5M5, Q8IWA2 | Gene names | HSPA4L, APG1, OSP94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
2A5B_HUMAN
|
||||||
| NC score | 0.007212 (rank : 22) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15173, Q13853 | Gene names | PPP2R5B | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit beta isoform (PP2A, B subunit, B' beta isoform) (PP2A, B subunit, B56 beta isoform) (PP2A, B subunit, PR61 beta isoform) (PP2A, B subunit, R5 beta isoform). | |||||
|
HSP74_MOUSE
|
||||||
| NC score | 0.007091 (rank : 23) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61316 | Gene names | Hspa4, Hsp110 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4 (Heat shock 70-related protein APG-2). | |||||
|
UB2D1_HUMAN
|
||||||
| NC score | 0.007004 (rank : 24) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51668 | Gene names | UBE2D1, UBCH5, UBCH5A | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 D1 (EC 6.3.2.19) (Ubiquitin-protein ligase D1) (Ubiquitin carrier protein D1) (UbcH5) (Ubiquitin- conjugating enzyme E2-17 kDa 1) (E2(17)KB 1). | |||||
|
UB2D1_MOUSE
|
||||||
| NC score | 0.007004 (rank : 25) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61080 | Gene names | Ube2d1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-conjugating enzyme E2 D1 (EC 6.3.2.19) (Ubiquitin-protein ligase D1) (Ubiquitin carrier protein D1) (Ubiquitin-conjugating enzyme E2-17 kDa 1) (E2(17)KB 1). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.006503 (rank : 26) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
SM1L2_MOUSE
|
||||||
| NC score | 0.000340 (rank : 27) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
LRRC4_HUMAN
|
||||||
| NC score | -0.000394 (rank : 28) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBW1, Q6ZMI8, Q96A85 | Gene names | LRRC4, BAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 4 precursor (Brain tumor- associated protein LRRC4) (NAG14). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | -0.000662 (rank : 29) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
CHK2_HUMAN
|
||||||
| NC score | -0.002508 (rank : 30) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O96017, Q6QA03, Q6QA04, Q6QA05, Q6QA06, Q6QA07, Q6QA08, Q6QA10, Q6QA11, Q6QA12, Q6QA13, Q9HCQ8, Q9UGF0, Q9UGF1 | Gene names | CHEK2, CHK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Chk2 (EC 2.7.11.1) (Cds1). | |||||