Please be patient as the page loads
|
TCAL8_MOUSE
|
||||||
| SwissProt Accessions | Q9CZY2, Q9CYP9 | Gene names | Tceal8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 8 (TCEA-like protein 8) (Transcription elongation factor S-II protein-like 8). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TCAL8_MOUSE
|
||||||
| θ value | 6.55044e-59 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9CZY2, Q9CYP9 | Gene names | Tceal8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 8 (TCEA-like protein 8) (Transcription elongation factor S-II protein-like 8). | |||||
|
TCAL8_HUMAN
|
||||||
| θ value | 2.42216e-37 (rank : 2) | NC score | 0.954951 (rank : 2) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IYN2 | Gene names | TCEAL8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 8 (TCEA-like protein 8) (Transcription elongation factor S-II protein-like 8). | |||||
|
TCAL7_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 3) | NC score | 0.821156 (rank : 3) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BRU2, Q96AT4 | Gene names | TCEAL7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 7 (TCEA-like protein 7) (Transcription elongation factor S-II protein-like 7). | |||||
|
TCAL1_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 4) | NC score | 0.647284 (rank : 4) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15170, Q9UJQ9 | Gene names | TCEAL1, SIIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 1 (TCEA-like protein 1) (Transcription elongation factor S-II protein-like 1) (Nuclear phosphoprotein p21/SIIR). | |||||
|
TCAL1_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 5) | NC score | 0.613716 (rank : 5) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q921P9 | Gene names | Tceal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 1 (TCEA-like protein 1) (Transcription elongation factor S-II protein-like 1). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 6) | NC score | 0.238752 (rank : 12) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TCAL3_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 7) | NC score | 0.328146 (rank : 7) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
TCAL6_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 8) | NC score | 0.342571 (rank : 6) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6IPX3, Q5H9J8 | Gene names | TCEAL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 6 (TCEA-like protein 6) (Transcription elongation factor S-II protein-like 6). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.094696 (rank : 13) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.023976 (rank : 33) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
TCAL5_MOUSE
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.307011 (rank : 8) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CCT4, Q497R6 | Gene names | Tceal5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TCAL3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.305535 (rank : 9) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R0A5, Q9CYK5, Q9D1S4 | Gene names | Tceal3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
MRP_MOUSE
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.088827 (rank : 14) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.052232 (rank : 20) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
PRRT3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.063620 (rank : 15) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
PRPC_HUMAN
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.058761 (rank : 17) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
UROL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.036131 (rank : 26) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5DID0, Q5DIC9, Q6LA40, Q6LA41, Q8N216 | Gene names | UMODL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin-like 1 precursor (Olfactorin). | |||||
|
ABCF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.032546 (rank : 27) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NE71, O14897, Q69YP6 | Gene names | ABCF1, ABC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein). | |||||
|
KBTBB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.022318 (rank : 34) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94819 | Gene names | KBTBD11, KIAA0711, KLHDC7C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch repeat and BTB domain-containing protein 11 (Kelch domain- containing protein 7B). | |||||
|
MAST1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.004168 (rank : 44) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
G3BP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.039844 (rank : 23) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UN86, O60606, O75149, Q9UPA1 | Gene names | G3BP2, KIAA0660 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.038261 (rank : 24) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.009832 (rank : 42) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
ARHG1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.019408 (rank : 36) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
TCAL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.276748 (rank : 10) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H3H9 | Gene names | TCEAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 2 (TCEA-like protein 2) (Transcription elongation factor S-II protein-like 2). | |||||
|
G3BP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.040297 (rank : 22) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97379, Q9R1B8 | Gene names | G3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.038087 (rank : 25) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PFTA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.027713 (rank : 30) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49354, Q9UDC1 | Gene names | FNTA | |||
|
Domain Architecture |
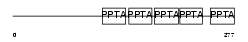
|
|||||
| Description | Protein farnesyltransferase/geranylgeranyltransferase type I alpha subunit (EC 2.5.1.58) (EC 2.5.1.59) (CAAX farnesyltransferase alpha subunit) (Ras proteins prenyltransferase alpha) (FTase-alpha) (Type I protein geranyl-geranyltransferase alpha subunit) (GGTase-I-alpha). | |||||
|
PHLB3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.027895 (rank : 29) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NSJ2, Q8N7Z4 | Gene names | PHLDB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 3. | |||||
|
PVRL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.025154 (rank : 32) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15223, O75465, Q9HBE6, Q9HBW2 | Gene names | PVRL1, HVEC, PRR1 | |||
|
Domain Architecture |

|
|||||
| Description | Poliovirus receptor-related protein 1 precursor (Herpes virus entry mediator C) (HveC) (Nectin-1) (Herpesvirus Ig-like receptor) (HIgR) (CD111 antigen). | |||||
|
PVRL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.026054 (rank : 31) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JKF6, Q9ERL5, Q9JI17 | Gene names | Pvrl1, Hvec, Prr1 | |||
|
Domain Architecture |

|
|||||
| Description | Poliovirus receptor-related protein 1 precursor (Herpes virus entry mediator C) (HveC) (Nectin-1) (CD111 antigen). | |||||
|
REXO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.031293 (rank : 28) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
CLD23_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.020564 (rank : 35) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96B33 | Gene names | CLDN23 | |||
|
Domain Architecture |
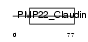
|
|||||
| Description | Claudin-23. | |||||
|
GA2L2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.016847 (rank : 37) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
PI52A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.014416 (rank : 39) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70172 | Gene names | Pip5k2a | |||
|
Domain Architecture |
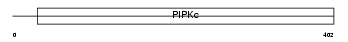
|
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-2 alpha (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type II alpha) (1- phosphatidylinositol-4-phosphate 5-kinase 2-alpha) (PtdIns(4)P-5- kinase isoform 2-alpha) (PIP5KII-alpha) (Diphosphoinositide kinase 2- alpha). | |||||
|
TCAL4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.263543 (rank : 11) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96EI5, Q8WY12, Q9H2H1, Q9H775 | Gene names | TCEAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 4 (TCEA-like protein 4) (Transcription elongation factor S-II protein-like 4). | |||||
|
CCD96_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.014911 (rank : 38) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 595 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q2M329, Q8N2I7 | Gene names | CCDC96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
FBX2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.011751 (rank : 41) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80UW2, Q8R0D2 | Gene names | Fbxo2, Fbx2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 2. | |||||
|
FGD5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.012899 (rank : 40) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.009321 (rank : 43) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
BEX5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.061416 (rank : 16) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5H9J7, Q569J0, Q56A74 | Gene names | NGFRAP1L1, BEX5, NADE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein BEX5 (Brain-expressed X-linked protein 5) (NGFRAP1-like 1) (Nerve growth factor receptor-associated protein 2). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.051718 (rank : 21) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.052948 (rank : 19) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.056339 (rank : 18) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
TCAL8_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 6.55044e-59 (rank : 1) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9CZY2, Q9CYP9 | Gene names | Tceal8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 8 (TCEA-like protein 8) (Transcription elongation factor S-II protein-like 8). | |||||
|
TCAL8_HUMAN
|
||||||
| NC score | 0.954951 (rank : 2) | θ value | 2.42216e-37 (rank : 2) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IYN2 | Gene names | TCEAL8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 8 (TCEA-like protein 8) (Transcription elongation factor S-II protein-like 8). | |||||
|
TCAL7_HUMAN
|
||||||
| NC score | 0.821156 (rank : 3) | θ value | 3.07829e-16 (rank : 3) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BRU2, Q96AT4 | Gene names | TCEAL7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 7 (TCEA-like protein 7) (Transcription elongation factor S-II protein-like 7). | |||||
|
TCAL1_HUMAN
|
||||||
| NC score | 0.647284 (rank : 4) | θ value | 3.64472e-09 (rank : 4) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15170, Q9UJQ9 | Gene names | TCEAL1, SIIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 1 (TCEA-like protein 1) (Transcription elongation factor S-II protein-like 1) (Nuclear phosphoprotein p21/SIIR). | |||||
|
TCAL1_MOUSE
|
||||||
| NC score | 0.613716 (rank : 5) | θ value | 2.36244e-08 (rank : 5) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q921P9 | Gene names | Tceal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 1 (TCEA-like protein 1) (Transcription elongation factor S-II protein-like 1). | |||||
|
TCAL6_HUMAN
|
||||||
| NC score | 0.342571 (rank : 6) | θ value | 0.00390308 (rank : 8) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6IPX3, Q5H9J8 | Gene names | TCEAL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 6 (TCEA-like protein 6) (Transcription elongation factor S-II protein-like 6). | |||||
|
TCAL3_HUMAN
|
||||||
| NC score | 0.328146 (rank : 7) | θ value | 0.000461057 (rank : 7) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
TCAL5_MOUSE
|
||||||
| NC score | 0.307011 (rank : 8) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CCT4, Q497R6 | Gene names | Tceal5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TCAL3_MOUSE
|
||||||
| NC score | 0.305535 (rank : 9) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R0A5, Q9CYK5, Q9D1S4 | Gene names | Tceal3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
TCAL2_HUMAN
|
||||||
| NC score | 0.276748 (rank : 10) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H3H9 | Gene names | TCEAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 2 (TCEA-like protein 2) (Transcription elongation factor S-II protein-like 2). | |||||
|
TCAL4_HUMAN
|
||||||
| NC score | 0.263543 (rank : 11) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96EI5, Q8WY12, Q9H2H1, Q9H775 | Gene names | TCEAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 4 (TCEA-like protein 4) (Transcription elongation factor S-II protein-like 4). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.238752 (rank : 12) | θ value | 0.000158464 (rank : 6) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.094696 (rank : 13) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
MRP_MOUSE
|
||||||
| NC score | 0.088827 (rank : 14) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
PRRT3_MOUSE
|
||||||
| NC score | 0.063620 (rank : 15) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
BEX5_HUMAN
|
||||||
| NC score | 0.061416 (rank : 16) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5H9J7, Q569J0, Q56A74 | Gene names | NGFRAP1L1, BEX5, NADE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein BEX5 (Brain-expressed X-linked protein 5) (NGFRAP1-like 1) (Nerve growth factor receptor-associated protein 2). | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.058761 (rank : 17) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PTMS_HUMAN
|
||||||
| NC score | 0.056339 (rank : 18) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.052948 (rank : 19) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.052232 (rank : 20) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.051718 (rank : 21) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
G3BP2_MOUSE
|
||||||
| NC score | 0.040297 (rank : 22) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97379, Q9R1B8 | Gene names | G3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
G3BP2_HUMAN
|
||||||
| NC score | 0.039844 (rank : 23) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UN86, O60606, O75149, Q9UPA1 | Gene names | G3BP2, KIAA0660 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.038261 (rank : 24) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.038087 (rank : 25) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
UROL1_HUMAN
|
||||||
| NC score | 0.036131 (rank : 26) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5DID0, Q5DIC9, Q6LA40, Q6LA41, Q8N216 | Gene names | UMODL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin-like 1 precursor (Olfactorin). | |||||
|
ABCF1_HUMAN
|
||||||
| NC score | 0.032546 (rank : 27) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NE71, O14897, Q69YP6 | Gene names | ABCF1, ABC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein). | |||||
|
REXO1_HUMAN
|
||||||
| NC score | 0.031293 (rank : 28) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
PHLB3_HUMAN
|
||||||
| NC score | 0.027895 (rank : 29) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NSJ2, Q8N7Z4 | Gene names | PHLDB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 3. | |||||
|
PFTA_HUMAN
|
||||||
| NC score | 0.027713 (rank : 30) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49354, Q9UDC1 | Gene names | FNTA | |||
|
Domain Architecture |

|
|||||
| Description | Protein farnesyltransferase/geranylgeranyltransferase type I alpha subunit (EC 2.5.1.58) (EC 2.5.1.59) (CAAX farnesyltransferase alpha subunit) (Ras proteins prenyltransferase alpha) (FTase-alpha) (Type I protein geranyl-geranyltransferase alpha subunit) (GGTase-I-alpha). | |||||
|
PVRL1_MOUSE
|
||||||
| NC score | 0.026054 (rank : 31) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JKF6, Q9ERL5, Q9JI17 | Gene names | Pvrl1, Hvec, Prr1 | |||
|
Domain Architecture |

|
|||||
| Description | Poliovirus receptor-related protein 1 precursor (Herpes virus entry mediator C) (HveC) (Nectin-1) (CD111 antigen). | |||||
|
PVRL1_HUMAN
|
||||||
| NC score | 0.025154 (rank : 32) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15223, O75465, Q9HBE6, Q9HBW2 | Gene names | PVRL1, HVEC, PRR1 | |||
|
Domain Architecture |

|
|||||
| Description | Poliovirus receptor-related protein 1 precursor (Herpes virus entry mediator C) (HveC) (Nectin-1) (Herpesvirus Ig-like receptor) (HIgR) (CD111 antigen). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.023976 (rank : 33) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
KBTBB_HUMAN
|
||||||
| NC score | 0.022318 (rank : 34) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94819 | Gene names | KBTBD11, KIAA0711, KLHDC7C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch repeat and BTB domain-containing protein 11 (Kelch domain- containing protein 7B). | |||||
|
CLD23_HUMAN
|
||||||
| NC score | 0.020564 (rank : 35) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96B33 | Gene names | CLDN23 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-23. | |||||
|
ARHG1_MOUSE
|
||||||
| NC score | 0.019408 (rank : 36) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
GA2L2_HUMAN
|
||||||
| NC score | 0.016847 (rank : 37) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
CCD96_HUMAN
|
||||||
| NC score | 0.014911 (rank : 38) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 595 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q2M329, Q8N2I7 | Gene names | CCDC96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
PI52A_MOUSE
|
||||||
| NC score | 0.014416 (rank : 39) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70172 | Gene names | Pip5k2a | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-2 alpha (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type II alpha) (1- phosphatidylinositol-4-phosphate 5-kinase 2-alpha) (PtdIns(4)P-5- kinase isoform 2-alpha) (PIP5KII-alpha) (Diphosphoinositide kinase 2- alpha). | |||||
|
FGD5_HUMAN
|
||||||
| NC score | 0.012899 (rank : 40) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZNL6, Q6MZY1, Q7Z303, Q8IYP3, Q8N861, Q8N8G4 | Gene names | FGD5, ZFYVE23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5 (Zinc finger FYVE domain-containing protein 23). | |||||
|
FBX2_MOUSE
|
||||||
| NC score | 0.011751 (rank : 41) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80UW2, Q8R0D2 | Gene names | Fbxo2, Fbx2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 2. | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.009832 (rank : 42) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.009321 (rank : 43) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
MAST1_MOUSE
|
||||||
| NC score | 0.004168 (rank : 44) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 40 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||