Please be patient as the page loads
|
SPEB_HUMAN
|
||||||
| SwissProt Accessions | Q9BSE5, Q5TDH1, Q9H5J3 | Gene names | AGMAT | |||
|
Domain Architecture |
|
|||||
| Description | Agmatinase, mitochondrial precursor (EC 3.5.3.11) (Agmatine ureohydrolase) (AUH). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SPEB_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BSE5, Q5TDH1, Q9H5J3 | Gene names | AGMAT | |||
|
Domain Architecture |

|
|||||
| Description | Agmatinase, mitochondrial precursor (EC 3.5.3.11) (Agmatine ureohydrolase) (AUH). | |||||
|
ARGI2_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 2) | NC score | 0.696418 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78540, Q6FHY8 | Gene names | ARG2 | |||
|
Domain Architecture |
|
|||||
| Description | Arginase-2, mitochondrial precursor (EC 3.5.3.1) (Arginase II) (Non- hepatic arginase) (Kidney-type arginase). | |||||
|
ARGI1_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 3) | NC score | 0.698522 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P05089, Q5JWT6, Q8TE72, Q9BS50 | Gene names | ARG1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginase-1 (EC 3.5.3.1) (Type I arginase) (Liver-type arginase). | |||||
|
ARGI1_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 4) | NC score | 0.688540 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61176, Q80VI4 | Gene names | Arg1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginase-1 (EC 3.5.3.1) (Type I arginase) (Liver-type arginase). | |||||
|
ARGI2_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 5) | NC score | 0.682046 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08691 | Gene names | Arg2 | |||
|
Domain Architecture |
|
|||||
| Description | Arginase-2, mitochondrial precursor (EC 3.5.3.1) (Arginase II) (Non- hepatic arginase) (Kidney-type arginase). | |||||
|
PYGO2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 6) | NC score | 0.054745 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BRQ0, Q8WYZ4, Q96CY2 | Gene names | PYGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 2. | |||||
|
CELR3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 7) | NC score | 0.003662 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
CPSF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 8) | NC score | 0.031100 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPU4 | Gene names | Cpsf1, Cpsf160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
CPSF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 9) | NC score | 0.029443 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q10570, Q96AF0 | Gene names | CPSF1, CPSF160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
HB21_HUMAN
|
||||||
| θ value | 8.99809 (rank : 10) | NC score | 0.009297 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01918, O19708, O19724, O78221, P01917 | Gene names | HLA-DQB, HLA-DRB1 | |||
|
Domain Architecture |
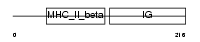
|
|||||
| Description | HLA class II histocompatibility antigen, DQ(1) beta chain precursor (DC-3 beta chain). | |||||
|
HB23_HUMAN
|
||||||
| θ value | 8.99809 (rank : 11) | NC score | 0.009267 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05537 | Gene names | ||||
|
Domain Architecture |
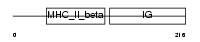
|
|||||
| Description | HLA class II histocompatibility antigen, DQ(W3) beta chain precursor. | |||||
|
HB24_HUMAN
|
||||||
| θ value | 8.99809 (rank : 12) | NC score | 0.009258 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01920 | Gene names | ||||
|
Domain Architecture |
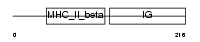
|
|||||
| Description | HLA class II histocompatibility antigen, DQ(3) beta chain precursor (Clone II-102). | |||||
|
SPEB_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BSE5, Q5TDH1, Q9H5J3 | Gene names | AGMAT | |||
|
Domain Architecture |

|
|||||
| Description | Agmatinase, mitochondrial precursor (EC 3.5.3.11) (Agmatine ureohydrolase) (AUH). | |||||
|
ARGI1_HUMAN
|
||||||
| NC score | 0.698522 (rank : 2) | θ value | 3.07829e-16 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P05089, Q5JWT6, Q8TE72, Q9BS50 | Gene names | ARG1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginase-1 (EC 3.5.3.1) (Type I arginase) (Liver-type arginase). | |||||
|
ARGI2_HUMAN
|
||||||
| NC score | 0.696418 (rank : 3) | θ value | 1.80466e-16 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78540, Q6FHY8 | Gene names | ARG2 | |||
|
Domain Architecture |

|
|||||
| Description | Arginase-2, mitochondrial precursor (EC 3.5.3.1) (Arginase II) (Non- hepatic arginase) (Kidney-type arginase). | |||||
|
ARGI1_MOUSE
|
||||||
| NC score | 0.688540 (rank : 4) | θ value | 2.20605e-14 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61176, Q80VI4 | Gene names | Arg1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginase-1 (EC 3.5.3.1) (Type I arginase) (Liver-type arginase). | |||||
|
ARGI2_MOUSE
|
||||||
| NC score | 0.682046 (rank : 5) | θ value | 1.09485e-13 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08691 | Gene names | Arg2 | |||
|
Domain Architecture |

|
|||||
| Description | Arginase-2, mitochondrial precursor (EC 3.5.3.1) (Arginase II) (Non- hepatic arginase) (Kidney-type arginase). | |||||
|
PYGO2_HUMAN
|
||||||
| NC score | 0.054745 (rank : 6) | θ value | 0.813845 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BRQ0, Q8WYZ4, Q96CY2 | Gene names | PYGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 2. | |||||
|
CPSF1_MOUSE
|
||||||
| NC score | 0.031100 (rank : 7) | θ value | 6.88961 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPU4 | Gene names | Cpsf1, Cpsf160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
CPSF1_HUMAN
|
||||||
| NC score | 0.029443 (rank : 8) | θ value | 8.99809 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q10570, Q96AF0 | Gene names | CPSF1, CPSF160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
HB21_HUMAN
|
||||||
| NC score | 0.009297 (rank : 9) | θ value | 8.99809 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01918, O19708, O19724, O78221, P01917 | Gene names | HLA-DQB, HLA-DRB1 | |||
|
Domain Architecture |

|
|||||
| Description | HLA class II histocompatibility antigen, DQ(1) beta chain precursor (DC-3 beta chain). | |||||
|
HB23_HUMAN
|
||||||
| NC score | 0.009267 (rank : 10) | θ value | 8.99809 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05537 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HLA class II histocompatibility antigen, DQ(W3) beta chain precursor. | |||||
|
HB24_HUMAN
|
||||||
| NC score | 0.009258 (rank : 11) | θ value | 8.99809 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01920 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HLA class II histocompatibility antigen, DQ(3) beta chain precursor (Clone II-102). | |||||
|
CELR3_HUMAN
|
||||||
| NC score | 0.003662 (rank : 12) | θ value | 6.88961 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||