Please be patient as the page loads
|
SP17_MOUSE
|
||||||
| SwissProt Accessions | Q62252 | Gene names | Spa17, Sp17 | |||
|
Domain Architecture |
|
|||||
| Description | Sperm surface protein Sp17 (Sperm autoantigenic protein 17). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SP17_MOUSE
|
||||||
| θ value | 8.55514e-59 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62252 | Gene names | Spa17, Sp17 | |||
|
Domain Architecture |

|
|||||
| Description | Sperm surface protein Sp17 (Sperm autoantigenic protein 17). | |||||
|
SP17_HUMAN
|
||||||
| θ value | 1.23823e-49 (rank : 2) | NC score | 0.972040 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15506, Q9BXF7 | Gene names | SPA17, SP17 | |||
|
Domain Architecture |
|
|||||
| Description | Sperm surface protein Sp17 (Sperm autoantigenic protein 17) (Sperm protein 17) (Sp17-1). | |||||
|
CABYR_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 3) | NC score | 0.394410 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75952, Q8WXW5, Q9HAY3, Q9HAY4, Q9HAY5, Q9HCY9 | Gene names | CABYR, CBP86, FSP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86) (Fibrousheathin-2) (FSP-2). | |||||
|
KAP2_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 4) | NC score | 0.240283 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
CABYR_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 5) | NC score | 0.359166 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D424, Q91Y41, Q91Y42 | Gene names | Cabyr, Cbp86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86). | |||||
|
NEMO_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 6) | NC score | 0.049757 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6K9 | Gene names | IKBKG, FIP3, NEMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (FIP-3). | |||||
|
KAP3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 7) | NC score | 0.202600 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP3_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.202009 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
MYO3A_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 9) | NC score | 0.016408 (rank : 29) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NEV4, Q8WX17, Q9NYS8 | Gene names | MYO3A | |||
|
Domain Architecture |

|
|||||
| Description | Myosin IIIA (EC 2.7.11.1). | |||||
|
KAP2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.193889 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
HIP1R_HUMAN
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.045612 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
SCN8A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 12) | NC score | 0.060062 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 1.06291 (rank : 13) | NC score | 0.040000 (rank : 26) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
NEUM_HUMAN
|
||||||
| θ value | 1.06291 (rank : 14) | NC score | 0.099405 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
SCN8A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 15) | NC score | 0.059669 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
STMN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.044772 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.050117 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
STMN1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.042647 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
CT151_HUMAN
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.040254 (rank : 25) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NC74, Q8N4Z9, Q9BR75, Q9H0Y9 | Gene names | C20orf151 | |||
|
Domain Architecture |
|
|||||
| Description | Uncharacterized protein C20orf151. | |||||
|
NEUG_HUMAN
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.147961 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92686 | Gene names | NRGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogranin (Ng) (RC3). | |||||
|
NEUG_MOUSE
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.142843 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P60761, Q3TYH4 | Gene names | Nrgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogranin (Ng) (RC3). | |||||
|
HNRPQ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.027426 (rank : 28) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60506, Q53H05, Q5TCG2, Q8IW78, Q8N599, Q96LC1, Q96LC2, Q9Y583 | Gene names | SYNCRIP, HNRPQ, NSAP1 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1). | |||||
|
HNRPQ_MOUSE
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.027542 (rank : 27) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TMK9, O88991, Q80YV6, Q8BGP1, Q8C5K6, Q8CGC2, Q91ZR0, Q99KU9, Q9QYF4 | Gene names | Syncrip, Hnrpq, Nsap1, Nsap1l | |||
|
Domain Architecture |
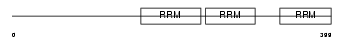
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1) (pp68). | |||||
|
SCN1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.045655 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
KAP0_HUMAN
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.102680 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
KAP0_MOUSE
|
||||||
| θ value | θ > 10 (rank : 26) | NC score | 0.103226 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
KAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 27) | NC score | 0.103161 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.103336 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
NEUM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.050861 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
SP17_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 8.55514e-59 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62252 | Gene names | Spa17, Sp17 | |||
|
Domain Architecture |

|
|||||
| Description | Sperm surface protein Sp17 (Sperm autoantigenic protein 17). | |||||
|
SP17_HUMAN
|
||||||
| NC score | 0.972040 (rank : 2) | θ value | 1.23823e-49 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15506, Q9BXF7 | Gene names | SPA17, SP17 | |||
|
Domain Architecture |

|
|||||
| Description | Sperm surface protein Sp17 (Sperm autoantigenic protein 17) (Sperm protein 17) (Sp17-1). | |||||
|
CABYR_HUMAN
|
||||||
| NC score | 0.394410 (rank : 3) | θ value | 1.29631e-06 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75952, Q8WXW5, Q9HAY3, Q9HAY4, Q9HAY5, Q9HCY9 | Gene names | CABYR, CBP86, FSP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86) (Fibrousheathin-2) (FSP-2). | |||||
|
CABYR_MOUSE
|
||||||
| NC score | 0.359166 (rank : 4) | θ value | 0.000461057 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D424, Q91Y41, Q91Y42 | Gene names | Cabyr, Cbp86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86). | |||||
|
KAP2_HUMAN
|
||||||
| NC score | 0.240283 (rank : 5) | θ value | 5.44631e-05 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP3_HUMAN
|
||||||
| NC score | 0.202600 (rank : 6) | θ value | 0.0431538 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP3_MOUSE
|
||||||
| NC score | 0.202009 (rank : 7) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP2_MOUSE
|
||||||
| NC score | 0.193889 (rank : 8) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
NEUG_HUMAN
|
||||||
| NC score | 0.147961 (rank : 9) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92686 | Gene names | NRGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogranin (Ng) (RC3). | |||||
|
NEUG_MOUSE
|
||||||
| NC score | 0.142843 (rank : 10) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P60761, Q3TYH4 | Gene names | Nrgn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogranin (Ng) (RC3). | |||||
|
KAP1_MOUSE
|
||||||
| NC score | 0.103336 (rank : 11) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP0_MOUSE
|
||||||
| NC score | 0.103226 (rank : 12) | θ value | θ > 10 (rank : 26) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
KAP1_HUMAN
|
||||||
| NC score | 0.103161 (rank : 13) | θ value | θ > 10 (rank : 27) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP0_HUMAN
|
||||||
| NC score | 0.102680 (rank : 14) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.099405 (rank : 15) | θ value | 1.06291 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
SCN8A_HUMAN
|
||||||
| NC score | 0.060062 (rank : 16) | θ value | 0.813845 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_MOUSE
|
||||||
| NC score | 0.059669 (rank : 17) | θ value | 1.06291 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.050861 (rank : 18) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.050117 (rank : 19) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
NEMO_HUMAN
|
||||||
| NC score | 0.049757 (rank : 20) | θ value | 0.0252991 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6K9 | Gene names | IKBKG, FIP3, NEMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (FIP-3). | |||||
|
SCN1A_HUMAN
|
||||||
| NC score | 0.045655 (rank : 21) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
HIP1R_HUMAN
|
||||||
| NC score | 0.045612 (rank : 22) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 809 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75146, Q9UED9 | Gene names | HIP1R, HIP12, KIAA0655 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related) (Hip 12). | |||||
|
STMN1_HUMAN
|
||||||
| NC score | 0.044772 (rank : 23) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
STMN1_MOUSE
|
||||||
| NC score | 0.042647 (rank : 24) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
CT151_HUMAN
|
||||||
| NC score | 0.040254 (rank : 25) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NC74, Q8N4Z9, Q9BR75, Q9H0Y9 | Gene names | C20orf151 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf151. | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.040000 (rank : 26) | θ value | 1.06291 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
HNRPQ_MOUSE
|
||||||
| NC score | 0.027542 (rank : 27) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TMK9, O88991, Q80YV6, Q8BGP1, Q8C5K6, Q8CGC2, Q91ZR0, Q99KU9, Q9QYF4 | Gene names | Syncrip, Hnrpq, Nsap1, Nsap1l | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1) (pp68). | |||||
|
HNRPQ_HUMAN
|
||||||
| NC score | 0.027426 (rank : 28) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60506, Q53H05, Q5TCG2, Q8IW78, Q8N599, Q96LC1, Q96LC2, Q9Y583 | Gene names | SYNCRIP, HNRPQ, NSAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein Q (hnRNP Q) (hnRNP-Q) (Synaptotagmin-binding, cytoplasmic RNA-interacting protein) (Glycine- and tyrosine-rich RNA-binding protein) (GRY-RBP) (NS1-associated protein 1). | |||||
|
MYO3A_HUMAN
|
||||||
| NC score | 0.016408 (rank : 29) | θ value | 0.0563607 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NEV4, Q8WX17, Q9NYS8 | Gene names | MYO3A | |||
|
Domain Architecture |

|
|||||
| Description | Myosin IIIA (EC 2.7.11.1). | |||||