Please be patient as the page loads
|
S10A6_HUMAN
|
||||||
| SwissProt Accessions | P06703 | Gene names | S100A6, CACY | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A6 (S100 calcium-binding protein A6) (Calcyclin) (Prolactin receptor-associated protein) (PRA) (Growth factor-inducible protein 2A9) (MLN 4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
S10A6_HUMAN
|
||||||
| θ value | 1.32909e-35 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P06703 | Gene names | S100A6, CACY | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A6 (S100 calcium-binding protein A6) (Calcyclin) (Prolactin receptor-associated protein) (PRA) (Growth factor-inducible protein 2A9) (MLN 4). | |||||
|
S10A6_MOUSE
|
||||||
| θ value | 1.24399e-33 (rank : 2) | NC score | 0.996799 (rank : 2) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P14069 | Gene names | S100a6, Cacy | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A6 (S100 calcium-binding protein A6) (Calcyclin) (Prolactin receptor-associated protein) (5B10). | |||||
|
S10A4_HUMAN
|
||||||
| θ value | 9.24701e-21 (rank : 3) | NC score | 0.919806 (rank : 6) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P26447 | Gene names | S100A4, CAPL, MTS1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A4 (S100 calcium-binding protein A4) (Metastasin) (Protein Mts1) (Placental calcium-binding protein) (Calvasculin). | |||||
|
S10A5_MOUSE
|
||||||
| θ value | 1.5773e-20 (rank : 4) | NC score | 0.930530 (rank : 4) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P63084, O88945, P82540 | Gene names | S100a5, S100d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein S100-A5 (S100 calcium-binding protein A5) (Protein S-100D). | |||||
|
S10A2_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 5) | NC score | 0.916353 (rank : 7) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P29034, O00266, Q9BU83 | Gene names | S100A2, S100L | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A2 (S100 calcium-binding protein A2) (Protein S-100L) (CAN19). | |||||
|
S10A4_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 6) | NC score | 0.920618 (rank : 5) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P07091, P20066 | Gene names | S100a4, Capl, Mts1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A4 (S100 calcium-binding protein A4) (Metastasin) (Protein Mts1) (Metastatic cell protein) (Placental calcium-binding protein) (18A2) (PEL98). | |||||
|
S10A1_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 7) | NC score | 0.877971 (rank : 8) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P56565, O88949 | Gene names | S100a1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A1 (S100 calcium-binding protein A1) (S-100 protein alpha subunit) (S-100 protein alpha chain). | |||||
|
S10A5_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 8) | NC score | 0.932369 (rank : 3) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P33763 | Gene names | S100A5, S100D | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A5 (S100 calcium-binding protein A5) (Protein S-100D). | |||||
|
S100Z_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 9) | NC score | 0.872226 (rank : 9) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WXG8 | Gene names | S100Z | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-Z (S100 calcium-binding protein Z). | |||||
|
S10AC_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 10) | NC score | 0.828063 (rank : 15) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P80511, P83219 | Gene names | S100A12 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A12 (S100 calcium-binding protein A12) (Calgranulin-C) (CAGC) (CGRP) (Neutrophil S100 protein) (Calcium-binding protein in amniotic fluid 1) (CAAF1) (p6) [Contains: Calcitermin]. | |||||
|
S10A9_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 11) | NC score | 0.837742 (rank : 14) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P31725 | Gene names | S100a9, Cagb, Mrp14 | |||
|
Domain Architecture |

|
|||||
| Description | Protein S100-A9 (S100 calcium-binding protein A9) (Calgranulin-B) (Migration inhibitory factor-related protein 14) (MRP-14) (P14) (Leukocyte L1 complex heavy chain). | |||||
|
S10AB_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 12) | NC score | 0.813090 (rank : 18) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P31949, Q5VTK0 | Gene names | S100A11, MLN70, S100C | |||
|
Domain Architecture |

|
|||||
| Description | Protein S100-A11 (S100 calcium-binding protein A11) (Protein S100C) (Calgizzarin) (MLN 70). | |||||
|
S10AB_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 13) | NC score | 0.822698 (rank : 17) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P50543, Q545S1 | Gene names | S100a11, S100c | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A11 (S100 calcium-binding protein A11) (Protein S100C) (Calgizzarin) (Endothelial monocyte-activating polypeptide) (EMAP). | |||||
|
S10A9_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 14) | NC score | 0.798695 (rank : 20) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P06702, Q9NYM0, Q9UCJ1 | Gene names | S100A9, CAGB, MRP14 | |||
|
Domain Architecture |

|
|||||
| Description | Protein S100-A9 (S100 calcium-binding protein A9) (Calgranulin-B) (Migration inhibitory factor-related protein 14) (MRP-14) (P14) (Leukocyte L1 complex heavy chain) (Calprotectin L1H subunit). | |||||
|
S10A3_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 15) | NC score | 0.824443 (rank : 16) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P62818, P56566 | Gene names | S100a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein S100-A3 (S100 calcium-binding protein A3) (Protein S-100E). | |||||
|
S100G_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 16) | NC score | 0.785281 (rank : 21) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P97816 | Gene names | S100g, Calb3, S100d | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-G (S100 calcium-binding protein G) (Vitamin D-dependent calcium-binding protein, intestinal) (CABP) (Calbindin D9K). | |||||
|
S100P_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 17) | NC score | 0.857734 (rank : 10) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P25815 | Gene names | S100P, S100E | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-P (S100 calcium-binding protein P). | |||||
|
S10A3_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 18) | NC score | 0.810565 (rank : 19) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P33764 | Gene names | S100A3, S100E | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A3 (S100 calcium-binding protein A3) (Protein S-100E). | |||||
|
S10A8_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 19) | NC score | 0.778050 (rank : 22) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27005, P31724 | Gene names | S100a8, Caga, Mrp8 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A8 (S100 calcium-binding protein A8) (Calgranulin-A) (Migration inhibitory factor-related protein 8) (MRP-8) (P8) (Leukocyte L1 complex light chain) (Chemotactic cytokine CP-10) (Pro- inflammatory S100 cytokine). | |||||
|
S10A1_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 20) | NC score | 0.857620 (rank : 11) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P23297 | Gene names | S100A1, S100A | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A1 (S100 calcium-binding protein A1) (S-100 protein alpha subunit) (S-100 protein alpha chain). | |||||
|
S100B_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 21) | NC score | 0.843589 (rank : 12) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P04271 | Gene names | S100B | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-B (S100 calcium-binding protein B) (S-100 protein beta subunit) (S-100 protein beta chain). | |||||
|
S100B_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 22) | NC score | 0.842412 (rank : 13) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P50114 | Gene names | S100b | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-B (S100 calcium-binding protein B) (S-100 protein beta subunit) (S-100 protein beta chain). | |||||
|
S100G_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 23) | NC score | 0.746281 (rank : 26) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P29377 | Gene names | S100G, CABP9K, CALB3, S100D | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-G (S100 calcium-binding protein G) (Vitamin D-dependent calcium-binding protein, intestinal) (CABP) (Calbindin D9K). | |||||
|
S10AD_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 24) | NC score | 0.740233 (rank : 28) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99584 | Gene names | S100A13 | |||
|
Domain Architecture |

|
|||||
| Description | Protein S100-A13 (S100 calcium-binding protein A13). | |||||
|
S11Y_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 25) | NC score | 0.743626 (rank : 27) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UDP3 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Putative S100 calcium-binding protein H_NH0456N16.1. | |||||
|
S10A8_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 26) | NC score | 0.762329 (rank : 24) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P05109, Q9UC92, Q9UCJ0 | Gene names | S100A8, CAGA, MRP8 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A8 (S100 calcium-binding protein A8) (Calgranulin-A) (Migration inhibitory factor-related protein 8) (MRP-8) (Cystic fibrosis antigen) (CFAG) (P8) (Leukocyte L1 complex light chain) (Calprotectin L1L subunit) (Urinary stone protein band A). | |||||
|
S10AA_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 27) | NC score | 0.764123 (rank : 23) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P60903, P08206 | Gene names | S100A10, ANX2LG, CAL1L, CLP11 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A10 (S100 calcium-binding protein A10) (Calpactin-1 light chain) (Calpactin I light chain) (p10 protein) (p11) (Cellular ligand of annexin II). | |||||
|
S10AA_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 28) | NC score | 0.758791 (rank : 25) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P08207 | Gene names | S100a10, Cal1l | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A10 (S100 calcium-binding protein A10) (Calpactin-1 light chain) (Calpactin I light chain) (p10 protein) (p11) (Cellular ligand of annexin II). | |||||
|
S10AD_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 29) | NC score | 0.730730 (rank : 30) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97352 | Gene names | S100a13 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A13 (S100 calcium-binding protein A13). | |||||
|
S10AG_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 30) | NC score | 0.735854 (rank : 29) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96FQ6 | Gene names | S100A16 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A16 (S100 calcium-binding protein A16) (Protein S100F). | |||||
|
HORN_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 31) | NC score | 0.449984 (rank : 33) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 32) | NC score | 0.125803 (rank : 44) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
SA7L1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 33) | NC score | 0.540207 (rank : 31) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86SG5 | Gene names | S100A7L1, S100A15 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A7-like 1 (S100 calcium-binding protein A7-like 1) (S100 calcium-binding protein A15). | |||||
|
CETN2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.102281 (rank : 56) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
RCN3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.057043 (rank : 85) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96D15, Q9HBZ8 | Gene names | RCN3 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulocalbin-3 precursor (EF-hand calcium-binding protein RLP49). | |||||
|
S10A7_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.470676 (rank : 32) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P31151, Q9H1E2 | Gene names | S100A7, PSOR1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A7 (S100 calcium-binding protein A7) (Psoriasin). | |||||
|
CABP5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.110991 (rank : 50) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CALN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.124040 (rank : 45) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BXU9 | Gene names | CALN1, CABP8 | |||
|
Domain Architecture |
|
|||||
| Description | Calneuron-1 (Calcium-binding protein CaBP8). | |||||
|
CALN1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.123997 (rank : 46) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JJG7 | Gene names | Caln1 | |||
|
Domain Architecture |
|
|||||
| Description | Calneuron-1. | |||||
|
CETN3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.097462 (rank : 62) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15182, Q9BS23 | Gene names | CETN3, CEN3 | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-3. | |||||
|
CETN3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.097447 (rank : 63) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35648 | Gene names | Cetn3, Cen3 | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-3. | |||||
|
CABP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.129897 (rank : 41) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
CABP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.129917 (rank : 40) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |
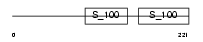
|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
CALM_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.092874 (rank : 64) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.092874 (rank : 65) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CETN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.101911 (rank : 57) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |
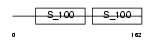
|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
CETN2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.100152 (rank : 58) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |
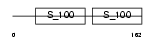
|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
RCN3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.050003 (rank : 101) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BH97, Q58E50, Q8R137, Q99K35, Q9CTD4 | Gene names | Rcn3, D7Ertd671e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulocalbin-3 precursor. | |||||
|
CABP7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.122057 (rank : 47) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q86V35 | Gene names | CABP7 | |||
|
Domain Architecture |
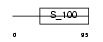
|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP7_MOUSE
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.122057 (rank : 48) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91ZM8 | Gene names | Cabp7 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CALL3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.087700 (rank : 67) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
CETN1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.100084 (rank : 59) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
S10AE_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.357829 (rank : 35) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D2Q8, Q9D1D8 | Gene names | S100a14, S100a15 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A14 (S100 calcium-binding protein A14) (S114). | |||||
|
TNNC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.107363 (rank : 52) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P02585 | Gene names | TNNC2 | |||
|
Domain Architecture |
|
|||||
| Description | Troponin C, skeletal muscle. | |||||
|
CABP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.128434 (rank : 42) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JLK4, Q9JLK5 | Gene names | Cabp2 | |||
|
Domain Architecture |
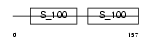
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CABP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.126249 (rank : 43) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NPB3 | Gene names | CABP2 | |||
|
Domain Architecture |
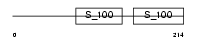
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
FKB10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.012310 (rank : 104) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96AY3, Q7Z3R4, Q9H3N3, Q9H6N5, Q9UF89 | Gene names | FKBP10, FKBP65 | |||
|
Domain Architecture |
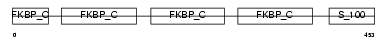
|
|||||
| Description | FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65). | |||||
|
TNNC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.105428 (rank : 54) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |
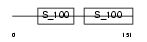
|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
ACTN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.026863 (rank : 102) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P35609, Q86TF4, Q86TI8 | Gene names | ACTN2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
ACTN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.025847 (rank : 103) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JI91 | Gene names | Actn2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
CABP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.105984 (rank : 53) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NZU6, Q9NYY0, Q9NZT9 | Gene names | CABP3 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 3 (CaBP3). | |||||
|
CABP5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.103882 (rank : 55) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CABP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.109909 (rank : 51) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CABP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.120835 (rank : 49) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CALL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.091524 (rank : 66) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CALL6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.083547 (rank : 69) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TD86, Q6Q2C4 | Gene names | CALML6, CAGLP, CALGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-like protein 6 (Calglandulin-like protein). | |||||
|
CALM4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.072800 (rank : 72) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
CANB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.083541 (rank : 70) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P63098, P06705, P15117, Q08044 | Gene names | PPP3R1, CNA2, CNB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.083549 (rank : 68) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q63810 | Gene names | Ppp3r1, Cnb | |||
|
Domain Architecture |
|
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.064851 (rank : 75) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96LZ3, Q7Z4V8, Q8WYJ4 | Gene names | PPP3R2, CBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform) (Calcineurin B-like protein) (CBLP) (CNBII). | |||||
|
CANB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.062200 (rank : 76) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |
|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
CAYP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.065934 (rank : 74) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13938, Q3MJA7, Q8WUZ3 | Gene names | CAPS | |||
|
Domain Architecture |
|
|||||
| Description | Calcyphosin (Calcyphosine). | |||||
|
EFCB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.071240 (rank : 73) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5VUJ9, Q59G23, Q9BS36 | Gene names | EFCAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
EFCB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.073425 (rank : 71) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9CQ46, Q9CS07 | Gene names | Efcab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
EFHD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.056589 (rank : 86) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BUP0, Q9BTF8, Q9H8I2, Q9HBQ0 | Gene names | EFHD1, SWS2 | |||
|
Domain Architecture |
|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
|
EFHD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.053174 (rank : 92) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D4J1, Q543U4 | Gene names | Efhd1, Sws2 | |||
|
Domain Architecture |
|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
|
FILA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.200565 (rank : 38) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
GPDM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.058464 (rank : 81) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P43304, Q59FR1, Q9HAP9 | Gene names | GPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M) (mtGPD). | |||||
|
HORN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.328506 (rank : 36) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
KIP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.052007 (rank : 95) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75838 | Gene names | CIB2, KIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
KIP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.051552 (rank : 97) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Z309 | Gene names | Cib2, Kip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
MLE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.053571 (rank : 91) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P05976 | Gene names | MYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
MLE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.052271 (rank : 94) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P05977 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
MLE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.053053 (rank : 93) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P06741 | Gene names | MYL1 | |||
|
Domain Architecture |
|
|||||
| Description | Myosin light chain 3, skeletal muscle isoform (A2 catalytic) (Alkali myosin light chain 3) (MLC3F). | |||||
|
MLE3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.051607 (rank : 96) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P05978 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |
|
|||||
| Description | Myosin light chain 3, skeletal muscle isoform (A2 catalytic) (Alkali myosin light chain 3) (MLC3F). | |||||
|
MLR5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.057517 (rank : 84) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02045 | Gene names | MYL5 | |||
|
Domain Architecture |
|
|||||
| Description | Superfast myosin regulatory light chain 2 (MyLC-2) (MYLC2) (Myosin regulatory light chain 5). | |||||
|
MLRA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.050832 (rank : 99) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QVP4, Q63977, Q9JIE8 | Gene names | Myl7, Mylc2a | |||
|
Domain Architecture |
|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
MLRM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.061349 (rank : 79) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P19105 | Gene names | MRLC2, MLCB, RLC | |||
|
Domain Architecture |
|
|||||
| Description | Myosin regulatory light chain 2, nonsarcomeric (Myosin RLC). | |||||
|
MLRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.061895 (rank : 77) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P24844, Q9H136 | Gene names | MYL9, MLC2, MRLC1, MYRL2 | |||
|
Domain Architecture |
|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9) (LC20). | |||||
|
MLRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.061837 (rank : 78) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9CQ19, Q80X77 | Gene names | Myl9, Myrl2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9). | |||||
|
MLRV_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.054714 (rank : 89) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P10916, Q16123 | Gene names | MYL2 | |||
|
Domain Architecture |
|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MLRV_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.054632 (rank : 90) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51667 | Gene names | Myl2, Mylpc | |||
|
Domain Architecture |
|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MYL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.051238 (rank : 98) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P09542, Q3UIF4, Q9CQZ2 | Gene names | Myl3, Mlc1v, Mylc | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain). | |||||
|
MYL6B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.058039 (rank : 82) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P14649 | Gene names | MYL6B, MLC1SA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B) (Myosin light chain 1 slow-twitch muscle A isoform) (MLC1sa). | |||||
|
MYL6B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.057783 (rank : 83) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CI43 | Gene names | Myl6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B). | |||||
|
MYL6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.055282 (rank : 87) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P60660, P16475, P24572, P24573, Q12790, Q6IAZ0, Q6IPY5 | Gene names | MYL6 | |||
|
Domain Architecture |
|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
MYL6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.055282 (rank : 88) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60605 | Gene names | Myl6, Myln | |||
|
Domain Architecture |
|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
PRVA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.050241 (rank : 100) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P20472, P78378, Q4VB78, Q5R3Q9 | Gene names | PVALB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parvalbumin alpha. | |||||
|
PRVA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.061044 (rank : 80) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P32848 | Gene names | Pvalb, Pva | |||
|
Domain Architecture |
|
|||||
| Description | Parvalbumin alpha. | |||||
|
RPTN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.240415 (rank : 37) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
RPTN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.140291 (rank : 39) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
S10AE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.383812 (rank : 34) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HCY8, Q5RHT0 | Gene names | S100A14, S100A15 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A14 (S100 calcium-binding protein A14) (S114). | |||||
|
TNNC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.098278 (rank : 61) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P63316, O14800, P02590, P04463 | Gene names | TNNC1, TNNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
TNNC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.098550 (rank : 60) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |
|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
S10A6_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.32909e-35 (rank : 1) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P06703 | Gene names | S100A6, CACY | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A6 (S100 calcium-binding protein A6) (Calcyclin) (Prolactin receptor-associated protein) (PRA) (Growth factor-inducible protein 2A9) (MLN 4). | |||||
|
S10A6_MOUSE
|
||||||
| NC score | 0.996799 (rank : 2) | θ value | 1.24399e-33 (rank : 2) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P14069 | Gene names | S100a6, Cacy | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A6 (S100 calcium-binding protein A6) (Calcyclin) (Prolactin receptor-associated protein) (5B10). | |||||
|
S10A5_HUMAN
|
||||||
| NC score | 0.932369 (rank : 3) | θ value | 2.28291e-11 (rank : 8) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P33763 | Gene names | S100A5, S100D | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A5 (S100 calcium-binding protein A5) (Protein S-100D). | |||||
|
S10A5_MOUSE
|
||||||
| NC score | 0.930530 (rank : 4) | θ value | 1.5773e-20 (rank : 4) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P63084, O88945, P82540 | Gene names | S100a5, S100d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein S100-A5 (S100 calcium-binding protein A5) (Protein S-100D). | |||||
|
S10A4_MOUSE
|
||||||
| NC score | 0.920618 (rank : 5) | θ value | 2.69671e-12 (rank : 6) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P07091, P20066 | Gene names | S100a4, Capl, Mts1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A4 (S100 calcium-binding protein A4) (Metastasin) (Protein Mts1) (Metastatic cell protein) (Placental calcium-binding protein) (18A2) (PEL98). | |||||
|
S10A4_HUMAN
|
||||||
| NC score | 0.919806 (rank : 6) | θ value | 9.24701e-21 (rank : 3) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P26447 | Gene names | S100A4, CAPL, MTS1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A4 (S100 calcium-binding protein A4) (Metastasin) (Protein Mts1) (Placental calcium-binding protein) (Calvasculin). | |||||
|
S10A2_HUMAN
|
||||||
| NC score | 0.916353 (rank : 7) | θ value | 3.63628e-17 (rank : 5) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P29034, O00266, Q9BU83 | Gene names | S100A2, S100L | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A2 (S100 calcium-binding protein A2) (Protein S-100L) (CAN19). | |||||
|
S10A1_MOUSE
|
||||||
| NC score | 0.877971 (rank : 8) | θ value | 1.02475e-11 (rank : 7) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P56565, O88949 | Gene names | S100a1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A1 (S100 calcium-binding protein A1) (S-100 protein alpha subunit) (S-100 protein alpha chain). | |||||
|
S100Z_HUMAN
|
||||||
| NC score | 0.872226 (rank : 9) | θ value | 2.98157e-11 (rank : 9) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WXG8 | Gene names | S100Z | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-Z (S100 calcium-binding protein Z). | |||||
|
S100P_HUMAN
|
||||||
| NC score | 0.857734 (rank : 10) | θ value | 2.21117e-06 (rank : 17) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P25815 | Gene names | S100P, S100E | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-P (S100 calcium-binding protein P). | |||||
|
S10A1_HUMAN
|
||||||
| NC score | 0.857620 (rank : 11) | θ value | 1.87187e-05 (rank : 20) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P23297 | Gene names | S100A1, S100A | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A1 (S100 calcium-binding protein A1) (S-100 protein alpha subunit) (S-100 protein alpha chain). | |||||
|
S100B_HUMAN
|
||||||
| NC score | 0.843589 (rank : 12) | θ value | 2.44474e-05 (rank : 21) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P04271 | Gene names | S100B | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-B (S100 calcium-binding protein B) (S-100 protein beta subunit) (S-100 protein beta chain). | |||||
|
S100B_MOUSE
|
||||||
| NC score | 0.842412 (rank : 13) | θ value | 2.44474e-05 (rank : 22) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P50114 | Gene names | S100b | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-B (S100 calcium-binding protein B) (S-100 protein beta subunit) (S-100 protein beta chain). | |||||
|
S10A9_MOUSE
|
||||||
| NC score | 0.837742 (rank : 14) | θ value | 1.38499e-08 (rank : 11) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P31725 | Gene names | S100a9, Cagb, Mrp14 | |||
|
Domain Architecture |

|
|||||
| Description | Protein S100-A9 (S100 calcium-binding protein A9) (Calgranulin-B) (Migration inhibitory factor-related protein 14) (MRP-14) (P14) (Leukocyte L1 complex heavy chain). | |||||
|
S10AC_HUMAN
|
||||||
| NC score | 0.828063 (rank : 15) | θ value | 6.21693e-09 (rank : 10) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P80511, P83219 | Gene names | S100A12 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A12 (S100 calcium-binding protein A12) (Calgranulin-C) (CAGC) (CGRP) (Neutrophil S100 protein) (Calcium-binding protein in amniotic fluid 1) (CAAF1) (p6) [Contains: Calcitermin]. | |||||
|
S10A3_MOUSE
|
||||||
| NC score | 0.824443 (rank : 16) | θ value | 5.81887e-07 (rank : 15) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P62818, P56566 | Gene names | S100a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein S100-A3 (S100 calcium-binding protein A3) (Protein S-100E). | |||||
|
S10AB_MOUSE
|
||||||
| NC score | 0.822698 (rank : 17) | θ value | 5.26297e-08 (rank : 13) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P50543, Q545S1 | Gene names | S100a11, S100c | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A11 (S100 calcium-binding protein A11) (Protein S100C) (Calgizzarin) (Endothelial monocyte-activating polypeptide) (EMAP). | |||||
|
S10AB_HUMAN
|
||||||
| NC score | 0.813090 (rank : 18) | θ value | 4.0297e-08 (rank : 12) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P31949, Q5VTK0 | Gene names | S100A11, MLN70, S100C | |||
|
Domain Architecture |

|
|||||
| Description | Protein S100-A11 (S100 calcium-binding protein A11) (Protein S100C) (Calgizzarin) (MLN 70). | |||||
|
S10A3_HUMAN
|
||||||
| NC score | 0.810565 (rank : 19) | θ value | 2.88788e-06 (rank : 18) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P33764 | Gene names | S100A3, S100E | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A3 (S100 calcium-binding protein A3) (Protein S-100E). | |||||
|
S10A9_HUMAN
|
||||||
| NC score | 0.798695 (rank : 20) | θ value | 3.41135e-07 (rank : 14) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P06702, Q9NYM0, Q9UCJ1 | Gene names | S100A9, CAGB, MRP14 | |||
|
Domain Architecture |

|
|||||
| Description | Protein S100-A9 (S100 calcium-binding protein A9) (Calgranulin-B) (Migration inhibitory factor-related protein 14) (MRP-14) (P14) (Leukocyte L1 complex heavy chain) (Calprotectin L1H subunit). | |||||
|
S100G_MOUSE
|
||||||
| NC score | 0.785281 (rank : 21) | θ value | 2.21117e-06 (rank : 16) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P97816 | Gene names | S100g, Calb3, S100d | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-G (S100 calcium-binding protein G) (Vitamin D-dependent calcium-binding protein, intestinal) (CABP) (Calbindin D9K). | |||||
|
S10A8_MOUSE
|
||||||
| NC score | 0.778050 (rank : 22) | θ value | 6.43352e-06 (rank : 19) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P27005, P31724 | Gene names | S100a8, Caga, Mrp8 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A8 (S100 calcium-binding protein A8) (Calgranulin-A) (Migration inhibitory factor-related protein 8) (MRP-8) (P8) (Leukocyte L1 complex light chain) (Chemotactic cytokine CP-10) (Pro- inflammatory S100 cytokine). | |||||
|
S10AA_HUMAN
|
||||||
| NC score | 0.764123 (rank : 23) | θ value | 9.29e-05 (rank : 27) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P60903, P08206 | Gene names | S100A10, ANX2LG, CAL1L, CLP11 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A10 (S100 calcium-binding protein A10) (Calpactin-1 light chain) (Calpactin I light chain) (p10 protein) (p11) (Cellular ligand of annexin II). | |||||
|
S10A8_HUMAN
|
||||||
| NC score | 0.762329 (rank : 24) | θ value | 9.29e-05 (rank : 26) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P05109, Q9UC92, Q9UCJ0 | Gene names | S100A8, CAGA, MRP8 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A8 (S100 calcium-binding protein A8) (Calgranulin-A) (Migration inhibitory factor-related protein 8) (MRP-8) (Cystic fibrosis antigen) (CFAG) (P8) (Leukocyte L1 complex light chain) (Calprotectin L1L subunit) (Urinary stone protein band A). | |||||
|
S10AA_MOUSE
|
||||||
| NC score | 0.758791 (rank : 25) | θ value | 0.00020696 (rank : 28) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P08207 | Gene names | S100a10, Cal1l | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A10 (S100 calcium-binding protein A10) (Calpactin-1 light chain) (Calpactin I light chain) (p10 protein) (p11) (Cellular ligand of annexin II). | |||||
|
S100G_HUMAN
|
||||||
| NC score | 0.746281 (rank : 26) | θ value | 3.19293e-05 (rank : 23) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P29377 | Gene names | S100G, CABP9K, CALB3, S100D | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-G (S100 calcium-binding protein G) (Vitamin D-dependent calcium-binding protein, intestinal) (CABP) (Calbindin D9K). | |||||
|
S11Y_HUMAN
|
||||||
| NC score | 0.743626 (rank : 27) | θ value | 5.44631e-05 (rank : 25) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UDP3 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Putative S100 calcium-binding protein H_NH0456N16.1. | |||||
|
S10AD_HUMAN
|
||||||
| NC score | 0.740233 (rank : 28) | θ value | 5.44631e-05 (rank : 24) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99584 | Gene names | S100A13 | |||
|
Domain Architecture |

|
|||||
| Description | Protein S100-A13 (S100 calcium-binding protein A13). | |||||
|
S10AG_HUMAN
|
||||||
| NC score | 0.735854 (rank : 29) | θ value | 0.00035302 (rank : 30) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96FQ6 | Gene names | S100A16 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A16 (S100 calcium-binding protein A16) (Protein S100F). | |||||
|
S10AD_MOUSE
|
||||||
| NC score | 0.730730 (rank : 30) | θ value | 0.00035302 (rank : 29) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97352 | Gene names | S100a13 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A13 (S100 calcium-binding protein A13). | |||||
|
SA7L1_HUMAN
|
||||||
| NC score | 0.540207 (rank : 31) | θ value | 0.0431538 (rank : 33) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86SG5 | Gene names | S100A7L1, S100A15 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A7-like 1 (S100 calcium-binding protein A7-like 1) (S100 calcium-binding protein A15). | |||||
|
S10A7_HUMAN
|
||||||
| NC score | 0.470676 (rank : 32) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P31151, Q9H1E2 | Gene names | S100A7, PSOR1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A7 (S100 calcium-binding protein A7) (Psoriasin). | |||||
|
HORN_MOUSE
|
||||||
| NC score | 0.449984 (rank : 33) | θ value | 0.00509761 (rank : 31) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
S10AE_HUMAN
|
||||||
| NC score | 0.383812 (rank : 34) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HCY8, Q5RHT0 | Gene names | S100A14, S100A15 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A14 (S100 calcium-binding protein A14) (S114). | |||||
|
S10AE_MOUSE
|
||||||
| NC score | 0.357829 (rank : 35) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D2Q8, Q9D1D8 | Gene names | S100a14, S100a15 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A14 (S100 calcium-binding protein A14) (S114). | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.328506 (rank : 36) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
RPTN_HUMAN
|
||||||
| NC score | 0.240415 (rank : 37) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.200565 (rank : 38) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.140291 (rank : 39) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
CABP1_MOUSE
|
||||||
| NC score | 0.129917 (rank : 40) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
CABP1_HUMAN
|
||||||
| NC score | 0.129897 (rank : 41) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
CABP2_MOUSE
|
||||||
| NC score | 0.128434 (rank : 42) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JLK4, Q9JLK5 | Gene names | Cabp2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CABP2_HUMAN
|
||||||
| NC score | 0.126249 (rank : 43) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NPB3 | Gene names | CABP2 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.125803 (rank : 44) | θ value | 0.00665767 (rank : 32) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
CALN1_HUMAN
|
||||||
| NC score | 0.124040 (rank : 45) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BXU9 | Gene names | CALN1, CABP8 | |||
|
Domain Architecture |

|
|||||
| Description | Calneuron-1 (Calcium-binding protein CaBP8). | |||||
|
CALN1_MOUSE
|
||||||
| NC score | 0.123997 (rank : 46) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JJG7 | Gene names | Caln1 | |||
|
Domain Architecture |

|
|||||
| Description | Calneuron-1. | |||||
|
CABP7_HUMAN
|
||||||
| NC score | 0.122057 (rank : 47) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q86V35 | Gene names | CABP7 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP7_MOUSE
|
||||||
| NC score | 0.122057 (rank : 48) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91ZM8 | Gene names | Cabp7 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP4_MOUSE
|
||||||
| NC score | 0.120835 (rank : 49) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CABP5_MOUSE
|
||||||
| NC score | 0.110991 (rank : 50) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CABP4_HUMAN
|
||||||
| NC score | 0.109909 (rank : 51) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
TNNC2_HUMAN
|
||||||
| NC score | 0.107363 (rank : 52) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P02585 | Gene names | TNNC2 | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, skeletal muscle. | |||||
|
CABP3_HUMAN
|
||||||
| NC score | 0.105984 (rank : 53) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NZU6, Q9NYY0, Q9NZT9 | Gene names | CABP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 3 (CaBP3). | |||||
|
TNNC2_MOUSE
|
||||||
| NC score | 0.105428 (rank : 54) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
CABP5_HUMAN
|
||||||
| NC score | 0.103882 (rank : 55) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CETN2_MOUSE
|
||||||
| NC score | 0.102281 (rank : 56) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CETN1_HUMAN
|
||||||
| NC score | 0.101911 (rank : 57) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
CETN2_HUMAN
|
||||||
| NC score | 0.100152 (rank : 58) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CETN1_MOUSE
|
||||||
| NC score | 0.100084 (rank : 59) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
TNNC1_MOUSE
|
||||||
| NC score | 0.098550 (rank : 60) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
TNNC1_HUMAN
|
||||||
| NC score | 0.098278 (rank : 61) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P63316, O14800, P02590, P04463 | Gene names | TNNC1, TNNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
CETN3_HUMAN
|
||||||
| NC score | 0.097462 (rank : 62) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15182, Q9BS23 | Gene names | CETN3, CEN3 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-3. | |||||
|
CETN3_MOUSE
|
||||||
| NC score | 0.097447 (rank : 63) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35648 | Gene names | Cetn3, Cen3 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-3. | |||||
|
CALM_HUMAN
|
||||||
| NC score | 0.092874 (rank : 64) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| NC score | 0.092874 (rank : 65) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALL5_HUMAN
|
||||||
| NC score | 0.091524 (rank : 66) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CALL3_HUMAN
|
||||||
| NC score | 0.087700 (rank : 67) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
CANB1_MOUSE
|
||||||
| NC score | 0.083549 (rank : 68) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q63810 | Gene names | Ppp3r1, Cnb | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CALL6_HUMAN
|
||||||
| NC score | 0.083547 (rank : 69) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TD86, Q6Q2C4 | Gene names | CALML6, CAGLP, CALGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-like protein 6 (Calglandulin-like protein). | |||||
|
CANB1_HUMAN
|
||||||
| NC score | 0.083541 (rank : 70) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P63098, P06705, P15117, Q08044 | Gene names | PPP3R1, CNA2, CNB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
EFCB2_MOUSE
|
||||||
| NC score | 0.073425 (rank : 71) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9CQ46, Q9CS07 | Gene names | Efcab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
CALM4_MOUSE
|
||||||
| NC score | 0.072800 (rank : 72) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
EFCB2_HUMAN
|
||||||
| NC score | 0.071240 (rank : 73) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5VUJ9, Q59G23, Q9BS36 | Gene names | EFCAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
CAYP1_HUMAN
|
||||||
| NC score | 0.065934 (rank : 74) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13938, Q3MJA7, Q8WUZ3 | Gene names | CAPS | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin (Calcyphosine). | |||||
|
CANB2_HUMAN
|
||||||
| NC score | 0.064851 (rank : 75) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96LZ3, Q7Z4V8, Q8WYJ4 | Gene names | PPP3R2, CBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform) (Calcineurin B-like protein) (CBLP) (CNBII). | |||||
|
CANB2_MOUSE
|
||||||
| NC score | 0.062200 (rank : 76) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
MLRN_HUMAN
|
||||||
| NC score | 0.061895 (rank : 77) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P24844, Q9H136 | Gene names | MYL9, MLC2, MRLC1, MYRL2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9) (LC20). | |||||
|
MLRN_MOUSE
|
||||||
| NC score | 0.061837 (rank : 78) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9CQ19, Q80X77 | Gene names | Myl9, Myrl2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9). | |||||
|
MLRM_HUMAN
|
||||||
| NC score | 0.061349 (rank : 79) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P19105 | Gene names | MRLC2, MLCB, RLC | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, nonsarcomeric (Myosin RLC). | |||||
|
PRVA_MOUSE
|
||||||
| NC score | 0.061044 (rank : 80) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P32848 | Gene names | Pvalb, Pva | |||
|
Domain Architecture |

|
|||||
| Description | Parvalbumin alpha. | |||||
|
GPDM_HUMAN
|
||||||
| NC score | 0.058464 (rank : 81) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P43304, Q59FR1, Q9HAP9 | Gene names | GPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M) (mtGPD). | |||||
|
MYL6B_HUMAN
|
||||||
| NC score | 0.058039 (rank : 82) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P14649 | Gene names | MYL6B, MLC1SA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B) (Myosin light chain 1 slow-twitch muscle A isoform) (MLC1sa). | |||||
|
MYL6B_MOUSE
|
||||||
| NC score | 0.057783 (rank : 83) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8CI43 | Gene names | Myl6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B). | |||||
|
MLR5_HUMAN
|
||||||
| NC score | 0.057517 (rank : 84) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02045 | Gene names | MYL5 | |||
|
Domain Architecture |

|
|||||
| Description | Superfast myosin regulatory light chain 2 (MyLC-2) (MYLC2) (Myosin regulatory light chain 5). | |||||
|
RCN3_HUMAN
|
||||||
| NC score | 0.057043 (rank : 85) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96D15, Q9HBZ8 | Gene names | RCN3 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulocalbin-3 precursor (EF-hand calcium-binding protein RLP49). | |||||
|
EFHD1_HUMAN
|
||||||
| NC score | 0.056589 (rank : 86) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BUP0, Q9BTF8, Q9H8I2, Q9HBQ0 | Gene names | EFHD1, SWS2 | |||
|
Domain Architecture |

|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
|
MYL6_HUMAN
|
||||||
| NC score | 0.055282 (rank : 87) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P60660, P16475, P24572, P24573, Q12790, Q6IAZ0, Q6IPY5 | Gene names | MYL6 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
MYL6_MOUSE
|
||||||
| NC score | 0.055282 (rank : 88) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60605 | Gene names | Myl6, Myln | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
MLRV_HUMAN
|
||||||
| NC score | 0.054714 (rank : 89) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P10916, Q16123 | Gene names | MYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MLRV_MOUSE
|
||||||
| NC score | 0.054632 (rank : 90) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51667 | Gene names | Myl2, Mylpc | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MLE1_HUMAN
|
||||||
| NC score | 0.053571 (rank : 91) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P05976 | Gene names | MYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
EFHD1_MOUSE
|
||||||
| NC score | 0.053174 (rank : 92) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D4J1, Q543U4 | Gene names | Efhd1, Sws2 | |||
|
Domain Architecture |

|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
|
MLE3_HUMAN
|
||||||
| NC score | 0.053053 (rank : 93) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P06741 | Gene names | MYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 3, skeletal muscle isoform (A2 catalytic) (Alkali myosin light chain 3) (MLC3F). | |||||
|
MLE1_MOUSE
|
||||||
| NC score | 0.052271 (rank : 94) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P05977 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
KIP2_HUMAN
|
||||||
| NC score | 0.052007 (rank : 95) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75838 | Gene names | CIB2, KIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
MLE3_MOUSE
|
||||||
| NC score | 0.051607 (rank : 96) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P05978 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 3, skeletal muscle isoform (A2 catalytic) (Alkali myosin light chain 3) (MLC3F). | |||||
|
KIP2_MOUSE
|
||||||
| NC score | 0.051552 (rank : 97) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Z309 | Gene names | Cib2, Kip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
MYL3_MOUSE
|
||||||
| NC score | 0.051238 (rank : 98) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P09542, Q3UIF4, Q9CQZ2 | Gene names | Myl3, Mlc1v, Mylc | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain). | |||||
|
MLRA_MOUSE
|
||||||
| NC score | 0.050832 (rank : 99) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QVP4, Q63977, Q9JIE8 | Gene names | Myl7, Mylc2a | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
PRVA_HUMAN
|
||||||
| NC score | 0.050241 (rank : 100) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P20472, P78378, Q4VB78, Q5R3Q9 | Gene names | PVALB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parvalbumin alpha. | |||||
|
RCN3_MOUSE
|
||||||
| NC score | 0.050003 (rank : 101) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BH97, Q58E50, Q8R137, Q99K35, Q9CTD4 | Gene names | Rcn3, D7Ertd671e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulocalbin-3 precursor. | |||||
|
ACTN2_HUMAN
|
||||||
| NC score | 0.026863 (rank : 102) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P35609, Q86TF4, Q86TI8 | Gene names | ACTN2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
ACTN2_MOUSE
|
||||||
| NC score | 0.025847 (rank : 103) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JI91 | Gene names | Actn2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
FKB10_HUMAN
|
||||||
| NC score | 0.012310 (rank : 104) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96AY3, Q7Z3R4, Q9H3N3, Q9H6N5, Q9UF89 | Gene names | FKBP10, FKBP65 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65). | |||||