Please be patient as the page loads
|
FKB10_HUMAN
|
||||||
| SwissProt Accessions | Q96AY3, Q7Z3R4, Q9H3N3, Q9H6N5, Q9UF89 | Gene names | FKBP10, FKBP65 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FKB10_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96AY3, Q7Z3R4, Q9H3N3, Q9H6N5, Q9UF89 | Gene names | FKBP10, FKBP65 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65). | |||||
|
FKB10_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997392 (rank : 2) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q61576, Q8VHI1 | Gene names | Fkbp10, Fkbp-rs, Fkbp1-rs, Fkbp6, Fkbprp | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65). | |||||
|
FKBP9_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.977671 (rank : 4) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95302, Q3MIR7, Q6IN76, Q6P2N1, Q96EX5, Q96IJ9 | Gene names | FKBP9, FKBP60, FKBP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 9 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase). | |||||
|
FKBP9_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.978183 (rank : 3) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Z247, Q80ZZ6, Q8R386, Q9CVM0, Q9JHX5 | Gene names | Fkbp9, Fkbp60, Fkbp63 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 9 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP65RS). | |||||
|
FKB14_HUMAN
|
||||||
| θ value | 1.42661e-21 (rank : 5) | NC score | 0.801349 (rank : 9) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NWM8 | Gene names | FKBP14, FKBP22 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 14 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (22 kDa FK506-binding protein) (FKBP-22). | |||||
|
FKBP2_HUMAN
|
||||||
| θ value | 7.0802e-21 (rank : 6) | NC score | 0.821419 (rank : 6) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26885, Q5BJH9, Q9BTS7 | Gene names | FKBP2, FKBP13 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
FKBP2_MOUSE
|
||||||
| θ value | 7.0802e-21 (rank : 7) | NC score | 0.825324 (rank : 5) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P45878 | Gene names | Fkbp2, Fkbp13 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
FKB14_MOUSE
|
||||||
| θ value | 9.24701e-21 (rank : 8) | NC score | 0.791376 (rank : 10) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P59024 | Gene names | Fkbp14 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 14 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase). | |||||
|
FKB11_MOUSE
|
||||||
| θ value | 6.62687e-19 (rank : 9) | NC score | 0.814169 (rank : 8) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D1M7, Q9CRE4 | Gene names | Fkbp11 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
FKB11_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 10) | NC score | 0.815397 (rank : 7) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NYL4 | Gene names | FKBP11, FKBP19 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
FKBP5_MOUSE
|
||||||
| θ value | 7.32683e-18 (rank : 11) | NC score | 0.594592 (rank : 19) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q64378 | Gene names | Fkbp5, Fkbp51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51). | |||||
|
FKBP7_MOUSE
|
||||||
| θ value | 1.63225e-17 (rank : 12) | NC score | 0.779104 (rank : 11) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O54998 | Gene names | Fkbp7, Fkbp23 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 7 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP-23). | |||||
|
FKBP5_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 13) | NC score | 0.583134 (rank : 20) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13451 | Gene names | FKBP5, AIG6, FKBP51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51) (54 kDa progesterone receptor-associated immunophilin) (FKBP54) (P54) (FF1 antigen) (HSP90-binding immunophilin) (Androgen-regulated protein 6). | |||||
|
FKB1A_HUMAN
|
||||||
| θ value | 7.58209e-15 (rank : 14) | NC score | 0.756755 (rank : 12) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P62942, P20071, Q6FGD9, Q6LEU3, Q9H103, Q9H566 | Gene names | FKBP1A, FKBP1, FKBP12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
FKBP4_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 15) | NC score | 0.582040 (rank : 21) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02790, Q9UCP1, Q9UCV7 | Gene names | FKBP4 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKB1A_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 16) | NC score | 0.750654 (rank : 13) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26883, Q545E9 | Gene names | Fkbp1a, Fkbp1 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
FKBP7_HUMAN
|
||||||
| θ value | 2.20605e-14 (rank : 17) | NC score | 0.735722 (rank : 16) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y680, Q86U65, Q96DA4, Q9Y6B0 | Gene names | FKBP7, FKBP23 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 7 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP-23). | |||||
|
FKBP4_MOUSE
|
||||||
| θ value | 8.38298e-14 (rank : 18) | NC score | 0.581077 (rank : 22) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P30416, Q3TVC9 | Gene names | Fkbp4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKB1B_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 19) | NC score | 0.749879 (rank : 15) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P68106, Q13664, Q16645, Q9BQ40 | Gene names | FKBP1B, FKBP12.6, FKBP1L, FKBP9, OTK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6) (h-FKBP-12). | |||||
|
FKB1B_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 20) | NC score | 0.750628 (rank : 14) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z2I2 | Gene names | Fkbp1b | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6). | |||||
|
FKBP3_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 21) | NC score | 0.687592 (rank : 18) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q00688, Q14317 | Gene names | FKBP3, FKBP25 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
FKBP3_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 22) | NC score | 0.689126 (rank : 17) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62446, Q9WTJ7 | Gene names | Fkbp3, Fkbp25 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
FKBP6_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 23) | NC score | 0.513660 (rank : 24) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75344, Q9UDS0 | Gene names | FKBP6, FKBP36 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
FKBP6_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 24) | NC score | 0.518192 (rank : 23) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91XW8, Q91Y30 | Gene names | Fkbp6 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
CSEN_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 25) | NC score | 0.147071 (rank : 28) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QXT8, Q924L0, Q99PH9, Q99PI0, Q99PI2, Q99PI3, Q9JHZ5 | Gene names | Kcnip3, Csen, Dream, Kchip3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (A-type potassium channel modulatory protein 3) (KChIP3). | |||||
|
KCIP1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 26) | NC score | 0.146530 (rank : 29) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZI2, Q5U822 | Gene names | KCNIP1, KCHIP1, VABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1) (Vesicle APC-binding protein). | |||||
|
KCIP1_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 27) | NC score | 0.146366 (rank : 30) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JJ57, Q5SSA3, Q6DTJ1, Q8BGJ4, Q8C4K4, Q8CGL1, Q8K1U1, Q8K3M2 | Gene names | Kcnip1, Kchip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1). | |||||
|
KCIP4_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 28) | NC score | 0.146021 (rank : 32) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6PIL6, Q4W5G8, Q8NEU0, Q9BWT2, Q9H294, Q9H2A4 | Gene names | KCNIP4, CALP, KCHIP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
KCIP4_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 29) | NC score | 0.146047 (rank : 31) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6PHZ8, Q6DTJ3, Q8CAD0, Q8R4I2, Q9EQ01 | Gene names | Kcnip4, Calp, Kchip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
CSEN_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 30) | NC score | 0.147984 (rank : 27) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y2W7, Q53TJ5, Q96T40, Q9UJ84, Q9UJ85 | Gene names | KCNIP3, CSEN, DREAM, KCHIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (KChIP3) (A-type potassium channel modulatory protein 3). | |||||
|
KCIP2_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 31) | NC score | 0.145648 (rank : 33) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JJ69, Q6DTJ2, Q8K1T8, Q8K1T9, Q8K1U0, Q8VHN4, Q8VHN5, Q8VHN6, Q9JJ68 | Gene names | Kcnip2, Kchip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2). | |||||
|
KCIP2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 32) | NC score | 0.144585 (rank : 34) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NS61, Q7Z6F1, Q96K86, Q96T41, Q96T42, Q96T43, Q96T44, Q9H0N4, Q9HD10, Q9HD11, Q9NS60, Q9NY10, Q9NZI1 | Gene names | KCNIP2, KCHIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2) (Cardiac voltage-gated potassium channel modulatory subunit). | |||||
|
FKBP8_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 33) | NC score | 0.321269 (rank : 25) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35465, Q99L93 | Gene names | Fkbp8, Fkbp38, Sam11 | |||
|
Domain Architecture |
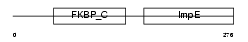
|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8) (muFKBP38). | |||||
|
VISL1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 34) | NC score | 0.112369 (rank : 39) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P62760, P28677, P29103, P42323, Q9UM20 | Gene names | VSNL1, VISL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Hippocalcin-like protein 3) (HLP3). | |||||
|
VISL1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 35) | NC score | 0.112369 (rank : 40) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P62761, P28677, P29103, P42323, Q9UM20 | Gene names | Vsnl1, Visl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Neural visinin-like protein 1) (NVL-1) (NVP-1). | |||||
|
TNNC1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 36) | NC score | 0.080333 (rank : 60) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P63316, O14800, P02590, P04463 | Gene names | TNNC1, TNNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
TNNC1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 37) | NC score | 0.080345 (rank : 59) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |
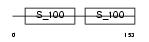
|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
NCS1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.114696 (rank : 35) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P62166, P36610, Q9UK26 | Gene names | FREQ, FLUP, NCS1 | |||
|
Domain Architecture |
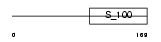
|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog) (Frequenin-like protein) (Frequenin-like ubiquitous protein). | |||||
|
NCS1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.114696 (rank : 36) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BNY6 | Gene names | Freq, Ncs1 | |||
|
Domain Architecture |
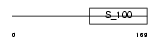
|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog). | |||||
|
FKBP8_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.303925 (rank : 26) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |
|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
KIP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.090212 (rank : 51) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z0F4, Q3TN80 | Gene names | Cib1, Cib, Kip, Prkdcip | |||
|
Domain Architecture |
|
|||||
| Description | Calcium and integrin-binding protein 1 (Calmyrin) (DNA-PKcs- interacting protein) (Kinase-interacting protein) (KIP) (CIB). | |||||
|
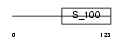
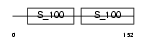
S100G_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.086129 (rank : 55) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97816 | Gene names | S100g, Calb3, S100d | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-G (S100 calcium-binding protein G) (Vitamin D-dependent calcium-binding protein, intestinal) (CABP) (Calbindin D9K). | |||||
|
TNNC2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.079401 (rank : 61) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |
|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
KIP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.089150 (rank : 54) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99828, O00693, O00735, Q6IB49, Q96J54, Q99971 | Gene names | CIB1, CIB, KIP, PRKDCIP | |||
|
Domain Architecture |
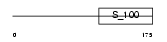
|
|||||
| Description | Calcium and integrin-binding protein 1 (Calmyrin) (DNA-PKcs- interacting protein) (Kinase-interacting protein) (KIP) (CIB) (SNK- interacting protein 2-28) (SIP2-28). | |||||
|
TNNC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.069499 (rank : 64) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P02585 | Gene names | TNNC2 | |||
|
Domain Architecture |
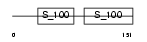
|
|||||
| Description | Troponin C, skeletal muscle. | |||||
|
HPCA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.112635 (rank : 37) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P84074, P32076, P41211, P70510 | Gene names | HPCA, BDR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin (Calcium-binding protein BDR-2). | |||||
|
HPCA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.112635 (rank : 38) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P84075, P32076, P41211, P70510 | Gene names | Hpca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin. | |||||
|
PLST_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.044808 (rank : 87) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P13797, Q86YI6 | Gene names | PLS3 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-3 (T-plastin). | |||||
|
CALU_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.040157 (rank : 90) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43852, O60456, Q6FHB9, Q96RL3, Q9NR43 | Gene names | CALU | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin) (IEF SSP 9302). | |||||
|
KDEL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.011425 (rank : 93) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z4H8, Q6UWW2, Q6ZUM9, Q8N7L8, Q8NE24 | Gene names | KDELC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KDEL motif-containing protein 2 precursor. | |||||
|
CALM4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.065955 (rank : 67) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
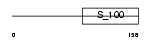
CAYP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.061746 (rank : 73) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13938, Q3MJA7, Q8WUZ3 | Gene names | CAPS | |||
|
Domain Architecture |
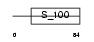
|
|||||
| Description | Calcyphosin (Calcyphosine). | |||||
|
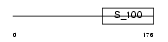
S10A6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.012310 (rank : 92) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06703 | Gene names | S100A6, CACY | |||
|
Domain Architecture |
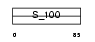
|
|||||
| Description | Protein S100-A6 (S100 calcium-binding protein A6) (Calcyclin) (Prolactin receptor-associated protein) (PRA) (Growth factor-inducible protein 2A9) (MLN 4). | |||||
|
CALB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.040462 (rank : 88) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
CALB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.040376 (rank : 89) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
CALU_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.038385 (rank : 91) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O35887 | Gene names | Calu | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin). | |||||
|
HPCL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.101727 (rank : 45) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P37235, Q969S5 | Gene names | HPCAL1, BDR1 | |||
|
Domain Architecture |
|
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Calcium-binding protein BDR-1) (HLP2). | |||||
|
HPCL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.101761 (rank : 44) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P62748, P35333 | Gene names | Hpcal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Neural visinin-like protein 3) (NVL-3) (NVP-3). | |||||
|
NCALD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.103511 (rank : 41) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P61601, P29554, Q8IYC3, Q9H0W2 | Gene names | NCALD | |||
|
Domain Architecture |
|
|||||
| Description | Neurocalcin delta. | |||||
|
NCALD_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.103511 (rank : 42) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91X97, Q3TJS9, Q8BZN9 | Gene names | Ncald, D15Ertd412e | |||
|
Domain Architecture |
|
|||||
| Description | Neurocalcin delta. | |||||
|
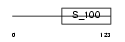
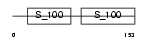
S100G_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.048709 (rank : 86) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P29377 | Gene names | S100G, CABP9K, CALB3, S100D | |||
|
Domain Architecture |
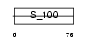
|
|||||
| Description | Protein S100-G (S100 calcium-binding protein G) (Vitamin D-dependent calcium-binding protein, intestinal) (CABP) (Calbindin D9K). | |||||
|
AIPL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.068010 (rank : 66) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIPL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.069136 (rank : 65) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q924K1 | Gene names | Aipl1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.081125 (rank : 57) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00170, Q99606 | Gene names | AIP, XAP2 | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein) (Immunophilin homolog ARA9) (HBV-X-associated protein 2). | |||||
|
AIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.080773 (rank : 58) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08915 | Gene names | Aip | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein). | |||||
|
CALL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.051204 (rank : 84) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CANB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.061670 (rank : 75) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P63098, P06705, P15117, Q08044 | Gene names | PPP3R1, CNA2, CNB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.061676 (rank : 74) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q63810 | Gene names | Ppp3r1, Cnb | |||
|
Domain Architecture |
|
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.057437 (rank : 77) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96LZ3, Q7Z4V8, Q8WYJ4 | Gene names | PPP3R2, CBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform) (Calcineurin B-like protein) (CBLP) (CNBII). | |||||
|
CANB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.058897 (rank : 76) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |
|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
CHP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.053273 (rank : 81) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43745 | Gene names | CHP2, HCA520 | |||
|
Domain Architecture |
|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520). | |||||
|
EFCB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.071667 (rank : 63) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HAE3 | Gene names | EFCAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
EFCB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.075213 (rank : 62) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D3N2, Q8BSW3, Q8C982, Q8CAI0, Q9D527, Q9D5E7 | Gene names | Efcab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
GUC1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.092777 (rank : 49) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P43080, Q9NU14 | Gene names | GUCA1A, GCAP, GCAP1, GUCA1 | |||
|
Domain Architecture |
|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
GUC1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.094535 (rank : 48) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P43081 | Gene names | Guca1a, Gcap, Gcap1, Guca1 | |||
|
Domain Architecture |
|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
GUC1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.091503 (rank : 50) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UMX6, Q9NU15 | Gene names | GUCA1B, GCAP2 | |||
|
Domain Architecture |
|
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
GUC1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.083642 (rank : 56) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VBV8 | Gene names | Guca1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
GUC1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.102222 (rank : 43) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95843, O95844, Q9UNM0 | Gene names | GUCA1C, GCAP3 | |||
|
Domain Architecture |
|
|||||
| Description | Guanylyl cyclase-activating protein 3 (GCAP 3) (Guanylate cyclase activator 1C). | |||||
|
HPCL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.096351 (rank : 47) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UM19, Q5TG97, Q8N611 | Gene names | HPCAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (HLP4). | |||||
|
HPCL4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.096352 (rank : 46) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BGZ1 | Gene names | Hpcal4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (Neural visinin-like protein 2) (NVP-2). | |||||
|
KIP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.050906 (rank : 85) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z309 | Gene names | Cib2, Kip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
OM34_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.065017 (rank : 69) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15785, Q9NTZ3 | Gene names | TOMM34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit) (hTom34). | |||||
|
OM34_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.065449 (rank : 68) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CYG7, Q3THD0, Q9D059 | Gene names | Tomm34 | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit). | |||||
|
PPID_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.061917 (rank : 71) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
PPID_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.063519 (rank : 70) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
RECO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.090167 (rank : 52) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35243 | Gene names | RCVRN, RCV1 | |||
|
Domain Architecture |
|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR). | |||||
|
RECO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.089830 (rank : 53) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P34057 | Gene names | Rcvrn, Rcv1 | |||
|
Domain Architecture |
|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR) (23 kDa photoreceptor cell-specific protein). | |||||
|
SPAG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.053132 (rank : 82) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
SPAG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.055952 (rank : 78) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
STUB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.054804 (rank : 79) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
STUB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.053364 (rank : 80) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
TTC12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.061882 (rank : 72) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
TTC12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.051427 (rank : 83) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BW49 | Gene names | Ttc12 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
FKB10_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96AY3, Q7Z3R4, Q9H3N3, Q9H6N5, Q9UF89 | Gene names | FKBP10, FKBP65 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65). | |||||
|
FKB10_MOUSE
|
||||||
| NC score | 0.997392 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q61576, Q8VHI1 | Gene names | Fkbp10, Fkbp-rs, Fkbp1-rs, Fkbp6, Fkbprp | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65). | |||||
|
FKBP9_MOUSE
|
||||||
| NC score | 0.978183 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Z247, Q80ZZ6, Q8R386, Q9CVM0, Q9JHX5 | Gene names | Fkbp9, Fkbp60, Fkbp63 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 9 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP65RS). | |||||
|
FKBP9_HUMAN
|
||||||
| NC score | 0.977671 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95302, Q3MIR7, Q6IN76, Q6P2N1, Q96EX5, Q96IJ9 | Gene names | FKBP9, FKBP60, FKBP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 9 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase). | |||||
|
FKBP2_MOUSE
|
||||||
| NC score | 0.825324 (rank : 5) | θ value | 7.0802e-21 (rank : 7) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P45878 | Gene names | Fkbp2, Fkbp13 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
FKBP2_HUMAN
|
||||||
| NC score | 0.821419 (rank : 6) | θ value | 7.0802e-21 (rank : 6) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26885, Q5BJH9, Q9BTS7 | Gene names | FKBP2, FKBP13 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
FKB11_HUMAN
|
||||||
| NC score | 0.815397 (rank : 7) | θ value | 2.5182e-18 (rank : 10) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NYL4 | Gene names | FKBP11, FKBP19 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
FKB11_MOUSE
|
||||||
| NC score | 0.814169 (rank : 8) | θ value | 6.62687e-19 (rank : 9) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D1M7, Q9CRE4 | Gene names | Fkbp11 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
FKB14_HUMAN
|
||||||
| NC score | 0.801349 (rank : 9) | θ value | 1.42661e-21 (rank : 5) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NWM8 | Gene names | FKBP14, FKBP22 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 14 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (22 kDa FK506-binding protein) (FKBP-22). | |||||
|
FKB14_MOUSE
|
||||||
| NC score | 0.791376 (rank : 10) | θ value | 9.24701e-21 (rank : 8) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P59024 | Gene names | Fkbp14 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 14 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase). | |||||
|
FKBP7_MOUSE
|
||||||
| NC score | 0.779104 (rank : 11) | θ value | 1.63225e-17 (rank : 12) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O54998 | Gene names | Fkbp7, Fkbp23 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 7 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP-23). | |||||
|
FKB1A_HUMAN
|
||||||
| NC score | 0.756755 (rank : 12) | θ value | 7.58209e-15 (rank : 14) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P62942, P20071, Q6FGD9, Q6LEU3, Q9H103, Q9H566 | Gene names | FKBP1A, FKBP1, FKBP12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
FKB1A_MOUSE
|
||||||
| NC score | 0.750654 (rank : 13) | θ value | 2.20605e-14 (rank : 16) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26883, Q545E9 | Gene names | Fkbp1a, Fkbp1 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
FKB1B_MOUSE
|
||||||
| NC score | 0.750628 (rank : 14) | θ value | 2.43908e-13 (rank : 20) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z2I2 | Gene names | Fkbp1b | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6). | |||||
|
FKB1B_HUMAN
|
||||||
| NC score | 0.749879 (rank : 15) | θ value | 2.43908e-13 (rank : 19) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P68106, Q13664, Q16645, Q9BQ40 | Gene names | FKBP1B, FKBP12.6, FKBP1L, FKBP9, OTK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6) (h-FKBP-12). | |||||
|
FKBP7_HUMAN
|
||||||
| NC score | 0.735722 (rank : 16) | θ value | 2.20605e-14 (rank : 17) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y680, Q86U65, Q96DA4, Q9Y6B0 | Gene names | FKBP7, FKBP23 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 7 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP-23). | |||||
|
FKBP3_MOUSE
|
||||||
| NC score | 0.689126 (rank : 17) | θ value | 2.52405e-10 (rank : 22) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62446, Q9WTJ7 | Gene names | Fkbp3, Fkbp25 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
FKBP3_HUMAN
|
||||||
| NC score | 0.687592 (rank : 18) | θ value | 1.47974e-10 (rank : 21) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q00688, Q14317 | Gene names | FKBP3, FKBP25 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
FKBP5_MOUSE
|
||||||
| NC score | 0.594592 (rank : 19) | θ value | 7.32683e-18 (rank : 11) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q64378 | Gene names | Fkbp5, Fkbp51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51). | |||||
|
FKBP5_HUMAN
|
||||||
| NC score | 0.583134 (rank : 20) | θ value | 1.80466e-16 (rank : 13) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13451 | Gene names | FKBP5, AIG6, FKBP51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51) (54 kDa progesterone receptor-associated immunophilin) (FKBP54) (P54) (FF1 antigen) (HSP90-binding immunophilin) (Androgen-regulated protein 6). | |||||
|
FKBP4_HUMAN
|
||||||
| NC score | 0.582040 (rank : 21) | θ value | 9.90251e-15 (rank : 15) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02790, Q9UCP1, Q9UCV7 | Gene names | FKBP4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP4_MOUSE
|
||||||
| NC score | 0.581077 (rank : 22) | θ value | 8.38298e-14 (rank : 18) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P30416, Q3TVC9 | Gene names | Fkbp4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP6_MOUSE
|
||||||
| NC score | 0.518192 (rank : 23) | θ value | 7.59969e-07 (rank : 24) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91XW8, Q91Y30 | Gene names | Fkbp6 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
FKBP6_HUMAN
|
||||||
| NC score | 0.513660 (rank : 24) | θ value | 1.53129e-07 (rank : 23) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75344, Q9UDS0 | Gene names | FKBP6, FKBP36 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
FKBP8_MOUSE
|
||||||
| NC score | 0.321269 (rank : 25) | θ value | 0.0736092 (rank : 33) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35465, Q99L93 | Gene names | Fkbp8, Fkbp38, Sam11 | |||
|
Domain Architecture |

|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8) (muFKBP38). | |||||
|
FKBP8_HUMAN
|
||||||
| NC score | 0.303925 (rank : 26) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |

|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
CSEN_HUMAN
|
||||||
| NC score | 0.147984 (rank : 27) | θ value | 0.00390308 (rank : 30) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y2W7, Q53TJ5, Q96T40, Q9UJ84, Q9UJ85 | Gene names | KCNIP3, CSEN, DREAM, KCHIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (KChIP3) (A-type potassium channel modulatory protein 3). | |||||
|
CSEN_MOUSE
|
||||||
| NC score | 0.147071 (rank : 28) | θ value | 0.00175202 (rank : 25) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QXT8, Q924L0, Q99PH9, Q99PI0, Q99PI2, Q99PI3, Q9JHZ5 | Gene names | Kcnip3, Csen, Dream, Kchip3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (A-type potassium channel modulatory protein 3) (KChIP3). | |||||
|
KCIP1_HUMAN
|
||||||
| NC score | 0.146530 (rank : 29) | θ value | 0.00228821 (rank : 26) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZI2, Q5U822 | Gene names | KCNIP1, KCHIP1, VABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1) (Vesicle APC-binding protein). | |||||
|
KCIP1_MOUSE
|
||||||
| NC score | 0.146366 (rank : 30) | θ value | 0.00228821 (rank : 27) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JJ57, Q5SSA3, Q6DTJ1, Q8BGJ4, Q8C4K4, Q8CGL1, Q8K1U1, Q8K3M2 | Gene names | Kcnip1, Kchip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1). | |||||
|
KCIP4_MOUSE
|
||||||
| NC score | 0.146047 (rank : 31) | θ value | 0.00298849 (rank : 29) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6PHZ8, Q6DTJ3, Q8CAD0, Q8R4I2, Q9EQ01 | Gene names | Kcnip4, Calp, Kchip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
KCIP4_HUMAN
|
||||||
| NC score | 0.146021 (rank : 32) | θ value | 0.00298849 (rank : 28) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6PIL6, Q4W5G8, Q8NEU0, Q9BWT2, Q9H294, Q9H2A4 | Gene names | KCNIP4, CALP, KCHIP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
KCIP2_MOUSE
|
||||||
| NC score | 0.145648 (rank : 33) | θ value | 0.00665767 (rank : 31) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JJ69, Q6DTJ2, Q8K1T8, Q8K1T9, Q8K1U0, Q8VHN4, Q8VHN5, Q8VHN6, Q9JJ68 | Gene names | Kcnip2, Kchip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2). | |||||
|
KCIP2_HUMAN
|
||||||
| NC score | 0.144585 (rank : 34) | θ value | 0.00869519 (rank : 32) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NS61, Q7Z6F1, Q96K86, Q96T41, Q96T42, Q96T43, Q96T44, Q9H0N4, Q9HD10, Q9HD11, Q9NS60, Q9NY10, Q9NZI1 | Gene names | KCNIP2, KCHIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2) (Cardiac voltage-gated potassium channel modulatory subunit). | |||||
|
NCS1_HUMAN
|
||||||
| NC score | 0.114696 (rank : 35) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P62166, P36610, Q9UK26 | Gene names | FREQ, FLUP, NCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog) (Frequenin-like protein) (Frequenin-like ubiquitous protein). | |||||
|
NCS1_MOUSE
|
||||||
| NC score | 0.114696 (rank : 36) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BNY6 | Gene names | Freq, Ncs1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog). | |||||
|
HPCA_HUMAN
|
||||||
| NC score | 0.112635 (rank : 37) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P84074, P32076, P41211, P70510 | Gene names | HPCA, BDR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin (Calcium-binding protein BDR-2). | |||||
|
HPCA_MOUSE
|
||||||
| NC score | 0.112635 (rank : 38) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P84075, P32076, P41211, P70510 | Gene names | Hpca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin. | |||||
|
VISL1_HUMAN
|
||||||
| NC score | 0.112369 (rank : 39) | θ value | 0.21417 (rank : 34) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P62760, P28677, P29103, P42323, Q9UM20 | Gene names | VSNL1, VISL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Hippocalcin-like protein 3) (HLP3). | |||||
|
VISL1_MOUSE
|
||||||
| NC score | 0.112369 (rank : 40) | θ value | 0.21417 (rank : 35) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P62761, P28677, P29103, P42323, Q9UM20 | Gene names | Vsnl1, Visl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Neural visinin-like protein 1) (NVL-1) (NVP-1). | |||||
|
NCALD_HUMAN
|
||||||
| NC score | 0.103511 (rank : 41) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P61601, P29554, Q8IYC3, Q9H0W2 | Gene names | NCALD | |||
|
Domain Architecture |

|
|||||
| Description | Neurocalcin delta. | |||||
|
NCALD_MOUSE
|
||||||
| NC score | 0.103511 (rank : 42) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91X97, Q3TJS9, Q8BZN9 | Gene names | Ncald, D15Ertd412e | |||
|
Domain Architecture |

|
|||||
| Description | Neurocalcin delta. | |||||
|
GUC1C_HUMAN
|
||||||
| NC score | 0.102222 (rank : 43) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95843, O95844, Q9UNM0 | Gene names | GUCA1C, GCAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 3 (GCAP 3) (Guanylate cyclase activator 1C). | |||||
|
HPCL1_MOUSE
|
||||||
| NC score | 0.101761 (rank : 44) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P62748, P35333 | Gene names | Hpcal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Neural visinin-like protein 3) (NVL-3) (NVP-3). | |||||
|
HPCL1_HUMAN
|
||||||
| NC score | 0.101727 (rank : 45) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P37235, Q969S5 | Gene names | HPCAL1, BDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Calcium-binding protein BDR-1) (HLP2). | |||||
|
HPCL4_MOUSE
|
||||||
| NC score | 0.096352 (rank : 46) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BGZ1 | Gene names | Hpcal4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (Neural visinin-like protein 2) (NVP-2). | |||||
|
HPCL4_HUMAN
|
||||||
| NC score | 0.096351 (rank : 47) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UM19, Q5TG97, Q8N611 | Gene names | HPCAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (HLP4). | |||||
|
GUC1A_MOUSE
|
||||||
| NC score | 0.094535 (rank : 48) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P43081 | Gene names | Guca1a, Gcap, Gcap1, Guca1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
GUC1A_HUMAN
|
||||||
| NC score | 0.092777 (rank : 49) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P43080, Q9NU14 | Gene names | GUCA1A, GCAP, GCAP1, GUCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
GUC1B_HUMAN
|
||||||
| NC score | 0.091503 (rank : 50) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UMX6, Q9NU15 | Gene names | GUCA1B, GCAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
KIP1_MOUSE
|
||||||
| NC score | 0.090212 (rank : 51) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z0F4, Q3TN80 | Gene names | Cib1, Cib, Kip, Prkdcip | |||
|
Domain Architecture |

|
|||||
| Description | Calcium and integrin-binding protein 1 (Calmyrin) (DNA-PKcs- interacting protein) (Kinase-interacting protein) (KIP) (CIB). | |||||
|
RECO_HUMAN
|
||||||
| NC score | 0.090167 (rank : 52) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35243 | Gene names | RCVRN, RCV1 | |||
|
Domain Architecture |

|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR). | |||||
|
RECO_MOUSE
|
||||||
| NC score | 0.089830 (rank : 53) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P34057 | Gene names | Rcvrn, Rcv1 | |||
|
Domain Architecture |

|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR) (23 kDa photoreceptor cell-specific protein). | |||||
|
KIP1_HUMAN
|
||||||
| NC score | 0.089150 (rank : 54) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99828, O00693, O00735, Q6IB49, Q96J54, Q99971 | Gene names | CIB1, CIB, KIP, PRKDCIP | |||
|
Domain Architecture |

|
|||||
| Description | Calcium and integrin-binding protein 1 (Calmyrin) (DNA-PKcs- interacting protein) (Kinase-interacting protein) (KIP) (CIB) (SNK- interacting protein 2-28) (SIP2-28). | |||||
|
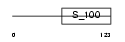
S100G_MOUSE
|
||||||
| NC score | 0.086129 (rank : 55) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97816 | Gene names | S100g, Calb3, S100d | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-G (S100 calcium-binding protein G) (Vitamin D-dependent calcium-binding protein, intestinal) (CABP) (Calbindin D9K). | |||||
|
GUC1B_MOUSE
|
||||||
| NC score | 0.083642 (rank : 56) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VBV8 | Gene names | Guca1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
AIP_HUMAN
|
||||||
| NC score | 0.081125 (rank : 57) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00170, Q99606 | Gene names | AIP, XAP2 | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein) (Immunophilin homolog ARA9) (HBV-X-associated protein 2). | |||||
|
AIP_MOUSE
|
||||||
| NC score | 0.080773 (rank : 58) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08915 | Gene names | Aip | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein). | |||||
|
TNNC1_MOUSE
|
||||||
| NC score | 0.080345 (rank : 59) | θ value | 0.279714 (rank : 37) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
TNNC1_HUMAN
|
||||||
| NC score | 0.080333 (rank : 60) | θ value | 0.279714 (rank : 36) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P63316, O14800, P02590, P04463 | Gene names | TNNC1, TNNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
TNNC2_MOUSE
|
||||||
| NC score | 0.079401 (rank : 61) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
EFCB1_MOUSE
|
||||||
| NC score | 0.075213 (rank : 62) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D3N2, Q8BSW3, Q8C982, Q8CAI0, Q9D527, Q9D5E7 | Gene names | Efcab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
EFCB1_HUMAN
|
||||||
| NC score | 0.071667 (rank : 63) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HAE3 | Gene names | EFCAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
TNNC2_HUMAN
|
||||||
| NC score | 0.069499 (rank : 64) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P02585 | Gene names | TNNC2 | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, skeletal muscle. | |||||
|
AIPL1_MOUSE
|
||||||
| NC score | 0.069136 (rank : 65) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q924K1 | Gene names | Aipl1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIPL1_HUMAN
|
||||||
| NC score | 0.068010 (rank : 66) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
CALM4_MOUSE
|
||||||
| NC score | 0.065955 (rank : 67) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
OM34_MOUSE
|
||||||
| NC score | 0.065449 (rank : 68) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CYG7, Q3THD0, Q9D059 | Gene names | Tomm34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit). | |||||
|
OM34_HUMAN
|
||||||
| NC score | 0.065017 (rank : 69) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15785, Q9NTZ3 | Gene names | TOMM34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit) (hTom34). | |||||
|
PPID_MOUSE
|
||||||
| NC score | 0.063519 (rank : 70) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
PPID_HUMAN
|
||||||
| NC score | 0.061917 (rank : 71) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
TTC12_HUMAN
|
||||||
| NC score | 0.061882 (rank : 72) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
CAYP1_HUMAN
|
||||||
| NC score | 0.061746 (rank : 73) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13938, Q3MJA7, Q8WUZ3 | Gene names | CAPS | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin (Calcyphosine). | |||||
|
CANB1_MOUSE
|
||||||
| NC score | 0.061676 (rank : 74) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q63810 | Gene names | Ppp3r1, Cnb | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB1_HUMAN
|
||||||
| NC score | 0.061670 (rank : 75) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P63098, P06705, P15117, Q08044 | Gene names | PPP3R1, CNA2, CNB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB2_MOUSE
|
||||||
| NC score | 0.058897 (rank : 76) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
CANB2_HUMAN
|
||||||
| NC score | 0.057437 (rank : 77) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96LZ3, Q7Z4V8, Q8WYJ4 | Gene names | PPP3R2, CBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform) (Calcineurin B-like protein) (CBLP) (CNBII). | |||||
|
SPAG1_MOUSE
|
||||||
| NC score | 0.055952 (rank : 78) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
STUB1_HUMAN
|
||||||
| NC score | 0.054804 (rank : 79) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
STUB1_MOUSE
|
||||||
| NC score | 0.053364 (rank : 80) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
CHP2_HUMAN
|
||||||
| NC score | 0.053273 (rank : 81) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43745 | Gene names | CHP2, HCA520 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520). | |||||
|
SPAG1_HUMAN
|
||||||
| NC score | 0.053132 (rank : 82) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
TTC12_MOUSE
|
||||||
| NC score | 0.051427 (rank : 83) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BW49 | Gene names | Ttc12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
CALL5_HUMAN
|
||||||
| NC score | 0.051204 (rank : 84) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
KIP2_MOUSE
|
||||||
| NC score | 0.050906 (rank : 85) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z309 | Gene names | Cib2, Kip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
S100G_HUMAN
|
||||||
| NC score | 0.048709 (rank : 86) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P29377 | Gene names | S100G, CABP9K, CALB3, S100D | |||
|
Domain Architecture |
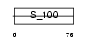
|
|||||
| Description | Protein S100-G (S100 calcium-binding protein G) (Vitamin D-dependent calcium-binding protein, intestinal) (CABP) (Calbindin D9K). | |||||
|
PLST_HUMAN
|
||||||
| NC score | 0.044808 (rank : 87) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P13797, Q86YI6 | Gene names | PLS3 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-3 (T-plastin). | |||||
|
CALB2_HUMAN
|
||||||
| NC score | 0.040462 (rank : 88) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
CALB2_MOUSE
|
||||||
| NC score | 0.040376 (rank : 89) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
CALU_HUMAN
|
||||||
| NC score | 0.040157 (rank : 90) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43852, O60456, Q6FHB9, Q96RL3, Q9NR43 | Gene names | CALU | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin) (IEF SSP 9302). | |||||
|
CALU_MOUSE
|
||||||
| NC score | 0.038385 (rank : 91) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O35887 | Gene names | Calu | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin). | |||||
|
S10A6_HUMAN
|
||||||
| NC score | 0.012310 (rank : 92) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06703 | Gene names | S100A6, CACY | |||
|
Domain Architecture |
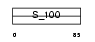
|
|||||
| Description | Protein S100-A6 (S100 calcium-binding protein A6) (Calcyclin) (Prolactin receptor-associated protein) (PRA) (Growth factor-inducible protein 2A9) (MLN 4). | |||||
|
KDEL2_HUMAN
|
||||||
| NC score | 0.011425 (rank : 93) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 61 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z4H8, Q6UWW2, Q6ZUM9, Q8N7L8, Q8NE24 | Gene names | KDELC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KDEL motif-containing protein 2 precursor. | |||||