Please be patient as the page loads
|
FKBP7_MOUSE
|
||||||
| SwissProt Accessions | O54998 | Gene names | Fkbp7, Fkbp23 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 7 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP-23). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FKBP7_MOUSE
|
||||||
| θ value | 1.13238e-127 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O54998 | Gene names | Fkbp7, Fkbp23 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 7 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP-23). | |||||
|
FKBP7_HUMAN
|
||||||
| θ value | 3.91911e-104 (rank : 2) | NC score | 0.979488 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y680, Q86U65, Q96DA4, Q9Y6B0 | Gene names | FKBP7, FKBP23 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 7 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP-23). | |||||
|
FKB14_MOUSE
|
||||||
| θ value | 4.70527e-49 (rank : 3) | NC score | 0.949488 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P59024 | Gene names | Fkbp14 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 14 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase). | |||||
|
FKB14_HUMAN
|
||||||
| θ value | 8.02596e-49 (rank : 4) | NC score | 0.945982 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NWM8 | Gene names | FKBP14, FKBP22 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 14 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (22 kDa FK506-binding protein) (FKBP-22). | |||||
|
FKBP9_HUMAN
|
||||||
| θ value | 1.09232e-21 (rank : 5) | NC score | 0.796416 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O95302, Q3MIR7, Q6IN76, Q6P2N1, Q96EX5, Q96IJ9 | Gene names | FKBP9, FKBP60, FKBP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 9 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase). | |||||
|
FKBP9_MOUSE
|
||||||
| θ value | 1.42661e-21 (rank : 6) | NC score | 0.796934 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Z247, Q80ZZ6, Q8R386, Q9CVM0, Q9JHX5 | Gene names | Fkbp9, Fkbp60, Fkbp63 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 9 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP65RS). | |||||
|
FKBP2_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 7) | NC score | 0.886077 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26885, Q5BJH9, Q9BTS7 | Gene names | FKBP2, FKBP13 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
FKBP2_MOUSE
|
||||||
| θ value | 2.69047e-20 (rank : 8) | NC score | 0.888790 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P45878 | Gene names | Fkbp2, Fkbp13 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
FKB10_MOUSE
|
||||||
| θ value | 6.62687e-19 (rank : 9) | NC score | 0.785392 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61576, Q8VHI1 | Gene names | Fkbp10, Fkbp-rs, Fkbp1-rs, Fkbp6, Fkbprp | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65). | |||||
|
FKB10_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 10) | NC score | 0.779104 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96AY3, Q7Z3R4, Q9H3N3, Q9H6N5, Q9UF89 | Gene names | FKBP10, FKBP65 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65). | |||||
|
FKB11_MOUSE
|
||||||
| θ value | 8.95645e-16 (rank : 11) | NC score | 0.836521 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D1M7, Q9CRE4 | Gene names | Fkbp11 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
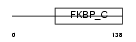
FKB11_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 12) | NC score | 0.832041 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NYL4 | Gene names | FKBP11, FKBP19 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
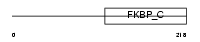
FKBP3_HUMAN
|
||||||
| θ value | 1.09485e-13 (rank : 13) | NC score | 0.778399 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q00688, Q14317 | Gene names | FKBP3, FKBP25 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
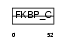
FKBP3_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 14) | NC score | 0.781473 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62446, Q9WTJ7 | Gene names | Fkbp3, Fkbp25 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
FKBP4_MOUSE
|
||||||
| θ value | 6.00763e-12 (rank : 15) | NC score | 0.607741 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P30416, Q3TVC9 | Gene names | Fkbp4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
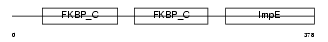
FKBP4_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 16) | NC score | 0.603215 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02790, Q9UCP1, Q9UCV7 | Gene names | FKBP4 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP5_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 17) | NC score | 0.602897 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13451 | Gene names | FKBP5, AIG6, FKBP51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51) (54 kDa progesterone receptor-associated immunophilin) (FKBP54) (P54) (FF1 antigen) (HSP90-binding immunophilin) (Androgen-regulated protein 6). | |||||
|
FKBP5_MOUSE
|
||||||
| θ value | 2.28291e-11 (rank : 18) | NC score | 0.609005 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q64378 | Gene names | Fkbp5, Fkbp51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51). | |||||
|
FKB1A_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 19) | NC score | 0.795340 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P62942, P20071, Q6FGD9, Q6LEU3, Q9H103, Q9H566 | Gene names | FKBP1A, FKBP1, FKBP12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
FKB1A_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 20) | NC score | 0.791859 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26883, Q545E9 | Gene names | Fkbp1a, Fkbp1 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
FKB1B_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 21) | NC score | 0.785292 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z2I2 | Gene names | Fkbp1b | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6). | |||||
|
FKB1B_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 22) | NC score | 0.781104 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P68106, Q13664, Q16645, Q9BQ40 | Gene names | FKBP1B, FKBP12.6, FKBP1L, FKBP9, OTK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6) (h-FKBP-12). | |||||
|
FKBP6_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 23) | NC score | 0.501929 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91XW8, Q91Y30 | Gene names | Fkbp6 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
FKBP6_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 24) | NC score | 0.489652 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75344, Q9UDS0 | Gene names | FKBP6, FKBP36 | |||
|
Domain Architecture |
|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
FKBP8_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 25) | NC score | 0.336359 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |
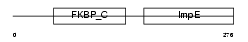
|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
FKBP8_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 26) | NC score | 0.351580 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35465, Q99L93 | Gene names | Fkbp8, Fkbp38, Sam11 | |||
|
Domain Architecture |
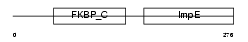
|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8) (muFKBP38). | |||||
|
RCN2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 27) | NC score | 0.126585 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BP92, O70341 | Gene names | Rcn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulocalbin-2 precursor (Taipoxin-associated calcium-binding protein 49) (TCBP-49). | |||||
|
CABP3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.072780 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZU6, Q9NYY0, Q9NZT9 | Gene names | CABP3 | |||
|
Domain Architecture |
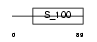
|
|||||
| Description | Calcium-binding protein 3 (CaBP3). | |||||
|
RCN2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.115902 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14257 | Gene names | RCN2, ERC55 | |||
|
Domain Architecture |
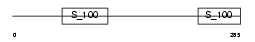
|
|||||
| Description | Reticulocalbin-2 precursor (Calcium-binding protein ERC-55) (E6- binding protein) (E6BP). | |||||
|
S100G_HUMAN
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.083257 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P29377 | Gene names | S100G, CABP9K, CALB3, S100D | |||
|
Domain Architecture |
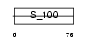
|
|||||
| Description | Protein S100-G (S100 calcium-binding protein G) (Vitamin D-dependent calcium-binding protein, intestinal) (CABP) (Calbindin D9K). | |||||
|
CALU_HUMAN
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.103366 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43852, O60456, Q6FHB9, Q96RL3, Q9NR43 | Gene names | CALU | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin) (IEF SSP 9302). | |||||
|
CALU_MOUSE
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.103359 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35887 | Gene names | Calu | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin). | |||||
|
S100G_MOUSE
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.100690 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97816 | Gene names | S100g, Calb3, S100d | |||
|
Domain Architecture |
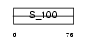
|
|||||
| Description | Protein S100-G (S100 calcium-binding protein G) (Vitamin D-dependent calcium-binding protein, intestinal) (CABP) (Calbindin D9K). | |||||
|
CABP5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.051175 (rank : 64) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
ONCO_HUMAN
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.079729 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P32930 | Gene names | OCM, OCMN | |||
|
Domain Architecture |
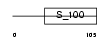
|
|||||
| Description | Oncomodulin (OM) (Parvalbumin beta). | |||||
|
CAB45_MOUSE
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.096256 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61112, Q61113 | Gene names | Sdf4, Cab45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 45 kDa calcium-binding protein precursor (Cab45) (Stromal cell-derived factor 4) (SDF-4). | |||||
|
CABP5_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.057150 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
ONCO_MOUSE
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.071185 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51879, Q62004 | Gene names | Ocm | |||
|
Domain Architecture |
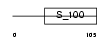
|
|||||
| Description | Oncomodulin (OM) (Parvalbumin beta). | |||||
|
RCN1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.094936 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q05186, Q3TVU3 | Gene names | Rcn1, Rca1, Rcn | |||
|
Domain Architecture |
|
|||||
| Description | Reticulocalbin-1 precursor. | |||||
|
CABP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.052686 (rank : 59) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
CABP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.052670 (rank : 60) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
DGKA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.022945 (rank : 74) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23743, O75481, O75482, O75483 | Gene names | DGKA, DAGK, DAGK1 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase alpha (EC 2.7.1.107) (Diglyceride kinase alpha) (DGK-alpha) (DAG kinase alpha) (80 kDa diacylglycerol kinase). | |||||
|
RCN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.101510 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15293 | Gene names | RCN1, RCN | |||
|
Domain Architecture |
|
|||||
| Description | Reticulocalbin-1 precursor. | |||||
|
NCALD_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.038897 (rank : 67) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P61601, P29554, Q8IYC3, Q9H0W2 | Gene names | NCALD | |||
|
Domain Architecture |
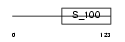
|
|||||
| Description | Neurocalcin delta. | |||||
|
NCALD_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.038897 (rank : 68) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91X97, Q3TJS9, Q8BZN9 | Gene names | Ncald, D15Ertd412e | |||
|
Domain Architecture |
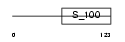
|
|||||
| Description | Neurocalcin delta. | |||||
|
CAB45_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.087924 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BRK5, Q8NBQ3, Q9NZP7 | Gene names | SDF4, CAB45 | |||
|
Domain Architecture |
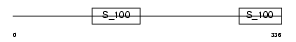
|
|||||
| Description | 45 kDa calcium-binding protein precursor (Cab45) (Stromal cell-derived factor 4) (SDF-4). | |||||
|
FSTL4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.008339 (rank : 78) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
S10A1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.022893 (rank : 75) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56565, O88949 | Gene names | S100a1 | |||
|
Domain Architecture |
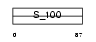
|
|||||
| Description | Protein S100-A1 (S100 calcium-binding protein A1) (S-100 protein alpha subunit) (S-100 protein alpha chain). | |||||
|
CALB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.040264 (rank : 65) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
CALB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.039839 (rank : 66) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
GLU2B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.008775 (rank : 77) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
HPCA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.037205 (rank : 69) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P84074, P32076, P41211, P70510 | Gene names | HPCA, BDR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin (Calcium-binding protein BDR-2). | |||||
|
HPCA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.037205 (rank : 70) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P84075, P32076, P41211, P70510 | Gene names | Hpca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin. | |||||
|
PLSI_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.033292 (rank : 71) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
RBY1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.004769 (rank : 79) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
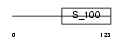
HPCL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.031847 (rank : 73) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P37235, Q969S5 | Gene names | HPCAL1, BDR1 | |||
|
Domain Architecture |
|
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Calcium-binding protein BDR-1) (HLP2). | |||||
|
HPCL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.032492 (rank : 72) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P62748, P35333 | Gene names | Hpcal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Neural visinin-like protein 3) (NVL-3) (NVP-3). | |||||
|
NUCB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.010441 (rank : 76) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P81117 | Gene names | Nucb2, Nefa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA). | |||||
|
AIPL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.076320 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIPL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.077831 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q924K1 | Gene names | Aipl1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.089215 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00170, Q99606 | Gene names | AIP, XAP2 | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein) (Immunophilin homolog ARA9) (HBV-X-associated protein 2). | |||||
|
AIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.088758 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08915 | Gene names | Aip | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein). | |||||
|
OM34_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.071371 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15785, Q9NTZ3 | Gene names | TOMM34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit) (hTom34). | |||||
|
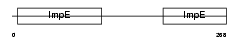
OM34_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.072048 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CYG7, Q3THD0, Q9D059 | Gene names | Tomm34 | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit). | |||||
|
PPID_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.069665 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
PPID_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.071396 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
RCN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.066951 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96D15, Q9HBZ8 | Gene names | RCN3 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulocalbin-3 precursor (EF-hand calcium-binding protein RLP49). | |||||
|
RCN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.070804 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BH97, Q58E50, Q8R137, Q99K35, Q9CTD4 | Gene names | Rcn3, D7Ertd671e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulocalbin-3 precursor. | |||||
|
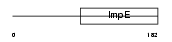
SGTB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.052583 (rank : 61) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96EQ0 | Gene names | SGTB, SGT2 | |||
|
Domain Architecture |
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B (Small glutamine-rich protein with tetratricopeptide repeats 2). | |||||
|
SGTB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.054758 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VD33 | Gene names | Sgtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B. | |||||
|
SPAG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.060766 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
SPAG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.063576 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
STUB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.059549 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
STUB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.057500 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
TTC12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.066914 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
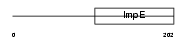
TTC12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.056285 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BW49 | Gene names | Ttc12 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
TTC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.051205 (rank : 63) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
TTC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.054484 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
Z3H7B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.051956 (rank : 62) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UGR2, Q5TFX9, Q8TBT9, Q9H8B6, Q9UGQ9, Q9UGR0, Q9UGR1, Q9UK03, Q9UPW9 | Gene names | ZC3H7B, KIAA1031 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7B (Rotavirus 'X'- associated non-structural protein) (RoXaN). | |||||
|
FKBP7_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.13238e-127 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O54998 | Gene names | Fkbp7, Fkbp23 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 7 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP-23). | |||||
|
FKBP7_HUMAN
|
||||||
| NC score | 0.979488 (rank : 2) | θ value | 3.91911e-104 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y680, Q86U65, Q96DA4, Q9Y6B0 | Gene names | FKBP7, FKBP23 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 7 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP-23). | |||||
|
FKB14_MOUSE
|
||||||
| NC score | 0.949488 (rank : 3) | θ value | 4.70527e-49 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P59024 | Gene names | Fkbp14 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 14 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase). | |||||
|
FKB14_HUMAN
|
||||||
| NC score | 0.945982 (rank : 4) | θ value | 8.02596e-49 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NWM8 | Gene names | FKBP14, FKBP22 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 14 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (22 kDa FK506-binding protein) (FKBP-22). | |||||
|
FKBP2_MOUSE
|
||||||
| NC score | 0.888790 (rank : 5) | θ value | 2.69047e-20 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P45878 | Gene names | Fkbp2, Fkbp13 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
FKBP2_HUMAN
|
||||||
| NC score | 0.886077 (rank : 6) | θ value | 4.15078e-21 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26885, Q5BJH9, Q9BTS7 | Gene names | FKBP2, FKBP13 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 2 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (13 kDa FKBP) (FKBP-13). | |||||
|
FKB11_MOUSE
|
||||||
| NC score | 0.836521 (rank : 7) | θ value | 8.95645e-16 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D1M7, Q9CRE4 | Gene names | Fkbp11 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
FKB11_HUMAN
|
||||||
| NC score | 0.832041 (rank : 8) | θ value | 3.40345e-15 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NYL4 | Gene names | FKBP11, FKBP19 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 11 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (19 kDa FK506-binding protein) (FKBP-19). | |||||
|
FKBP9_MOUSE
|
||||||
| NC score | 0.796934 (rank : 9) | θ value | 1.42661e-21 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Z247, Q80ZZ6, Q8R386, Q9CVM0, Q9JHX5 | Gene names | Fkbp9, Fkbp60, Fkbp63 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 9 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (FKBP65RS). | |||||
|
FKBP9_HUMAN
|
||||||
| NC score | 0.796416 (rank : 10) | θ value | 1.09232e-21 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O95302, Q3MIR7, Q6IN76, Q6P2N1, Q96EX5, Q96IJ9 | Gene names | FKBP9, FKBP60, FKBP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 9 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase). | |||||
|
FKB1A_HUMAN
|
||||||
| NC score | 0.795340 (rank : 11) | θ value | 1.133e-10 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P62942, P20071, Q6FGD9, Q6LEU3, Q9H103, Q9H566 | Gene names | FKBP1A, FKBP1, FKBP12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
FKB1A_MOUSE
|
||||||
| NC score | 0.791859 (rank : 12) | θ value | 3.29651e-10 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26883, Q545E9 | Gene names | Fkbp1a, Fkbp1 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 1A (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (12 kDa FKBP) (FKBP-12) (Immunophilin FKBP12). | |||||
|
FKB10_MOUSE
|
||||||
| NC score | 0.785392 (rank : 13) | θ value | 6.62687e-19 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61576, Q8VHI1 | Gene names | Fkbp10, Fkbp-rs, Fkbp1-rs, Fkbp6, Fkbprp | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65). | |||||
|
FKB1B_MOUSE
|
||||||
| NC score | 0.785292 (rank : 14) | θ value | 3.29651e-10 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z2I2 | Gene names | Fkbp1b | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6). | |||||
|
FKBP3_MOUSE
|
||||||
| NC score | 0.781473 (rank : 15) | θ value | 1.86753e-13 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62446, Q9WTJ7 | Gene names | Fkbp3, Fkbp25 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
FKB1B_HUMAN
|
||||||
| NC score | 0.781104 (rank : 16) | θ value | 9.59137e-10 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P68106, Q13664, Q16645, Q9BQ40 | Gene names | FKBP1B, FKBP12.6, FKBP1L, FKBP9, OTK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FK506-binding protein 1B (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase 1B) (PPIase 1B) (Rotamase 1B) (12.6 kDa FKBP) (FKBP-12.6) (Immunophilin FKBP12.6) (h-FKBP-12). | |||||
|
FKB10_HUMAN
|
||||||
| NC score | 0.779104 (rank : 17) | θ value | 1.63225e-17 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96AY3, Q7Z3R4, Q9H3N3, Q9H6N5, Q9UF89 | Gene names | FKBP10, FKBP65 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 10 precursor (EC 5.2.1.8) (Peptidyl-prolyl cis- trans isomerase) (PPIase) (Rotamase) (65 kDa FK506-binding protein) (FKBP65) (Immunophilin FKBP65). | |||||
|
FKBP3_HUMAN
|
||||||
| NC score | 0.778399 (rank : 18) | θ value | 1.09485e-13 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q00688, Q14317 | Gene names | FKBP3, FKBP25 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 3 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (25 kDa FKBP) (FKBP-25) (Rapamycin- selective 25 kDa immunophilin). | |||||
|
FKBP5_MOUSE
|
||||||
| NC score | 0.609005 (rank : 19) | θ value | 2.28291e-11 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q64378 | Gene names | Fkbp5, Fkbp51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51). | |||||
|
FKBP4_MOUSE
|
||||||
| NC score | 0.607741 (rank : 20) | θ value | 6.00763e-12 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P30416, Q3TVC9 | Gene names | Fkbp4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP4_HUMAN
|
||||||
| NC score | 0.603215 (rank : 21) | θ value | 2.28291e-11 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02790, Q9UCP1, Q9UCV7 | Gene names | FKBP4 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 4 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (p59 protein) (HSP-binding immunophilin) (HBI) (FKBP52 protein) (52 kDa FK506-binding protein) (FKBP59). | |||||
|
FKBP5_HUMAN
|
||||||
| NC score | 0.602897 (rank : 22) | θ value | 2.28291e-11 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13451 | Gene names | FKBP5, AIG6, FKBP51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51) (54 kDa progesterone receptor-associated immunophilin) (FKBP54) (P54) (FF1 antigen) (HSP90-binding immunophilin) (Androgen-regulated protein 6). | |||||
|
FKBP6_MOUSE
|
||||||
| NC score | 0.501929 (rank : 23) | θ value | 3.19293e-05 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91XW8, Q91Y30 | Gene names | Fkbp6 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
FKBP6_HUMAN
|
||||||
| NC score | 0.489652 (rank : 24) | θ value | 0.000158464 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75344, Q9UDS0 | Gene names | FKBP6, FKBP36 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 6 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (36 kDa FK506-binding protein) (FKBP- 36) (Immunophilin FKBP36). | |||||
|
FKBP8_MOUSE
|
||||||
| NC score | 0.351580 (rank : 25) | θ value | 0.0252991 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35465, Q99L93 | Gene names | Fkbp8, Fkbp38, Sam11 | |||
|
Domain Architecture |

|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8) (muFKBP38). | |||||
|
FKBP8_HUMAN
|
||||||
| NC score | 0.336359 (rank : 26) | θ value | 0.0193708 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14318 | Gene names | FKBP8, FKBP38 | |||
|
Domain Architecture |

|
|||||
| Description | 38 kDa FK506-binding protein homolog (FKBPR38) (FK506-binding protein 8). | |||||
|
RCN2_MOUSE
|
||||||
| NC score | 0.126585 (rank : 27) | θ value | 0.0431538 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BP92, O70341 | Gene names | Rcn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulocalbin-2 precursor (Taipoxin-associated calcium-binding protein 49) (TCBP-49). | |||||
|
RCN2_HUMAN
|
||||||
| NC score | 0.115902 (rank : 28) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14257 | Gene names | RCN2, ERC55 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulocalbin-2 precursor (Calcium-binding protein ERC-55) (E6- binding protein) (E6BP). | |||||
|
CALU_HUMAN
|
||||||
| NC score | 0.103366 (rank : 29) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43852, O60456, Q6FHB9, Q96RL3, Q9NR43 | Gene names | CALU | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin) (IEF SSP 9302). | |||||
|
CALU_MOUSE
|
||||||
| NC score | 0.103359 (rank : 30) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35887 | Gene names | Calu | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin). | |||||
|
RCN1_HUMAN
|
||||||
| NC score | 0.101510 (rank : 31) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15293 | Gene names | RCN1, RCN | |||
|
Domain Architecture |

|
|||||
| Description | Reticulocalbin-1 precursor. | |||||
|
S100G_MOUSE
|
||||||
| NC score | 0.100690 (rank : 32) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97816 | Gene names | S100g, Calb3, S100d | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-G (S100 calcium-binding protein G) (Vitamin D-dependent calcium-binding protein, intestinal) (CABP) (Calbindin D9K). | |||||
|
CAB45_MOUSE
|
||||||
| NC score | 0.096256 (rank : 33) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61112, Q61113 | Gene names | Sdf4, Cab45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 45 kDa calcium-binding protein precursor (Cab45) (Stromal cell-derived factor 4) (SDF-4). | |||||
|
RCN1_MOUSE
|
||||||
| NC score | 0.094936 (rank : 34) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q05186, Q3TVU3 | Gene names | Rcn1, Rca1, Rcn | |||
|
Domain Architecture |

|
|||||
| Description | Reticulocalbin-1 precursor. | |||||
|
AIP_HUMAN
|
||||||
| NC score | 0.089215 (rank : 35) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00170, Q99606 | Gene names | AIP, XAP2 | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein) (Immunophilin homolog ARA9) (HBV-X-associated protein 2). | |||||
|
AIP_MOUSE
|
||||||
| NC score | 0.088758 (rank : 36) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08915 | Gene names | Aip | |||
|
Domain Architecture |

|
|||||
| Description | AH receptor-interacting protein (AIP) (Aryl-hydrocarbon receptor- interacting protein). | |||||
|
CAB45_HUMAN
|
||||||
| NC score | 0.087924 (rank : 37) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BRK5, Q8NBQ3, Q9NZP7 | Gene names | SDF4, CAB45 | |||
|
Domain Architecture |

|
|||||
| Description | 45 kDa calcium-binding protein precursor (Cab45) (Stromal cell-derived factor 4) (SDF-4). | |||||
|
S100G_HUMAN
|
||||||
| NC score | 0.083257 (rank : 38) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P29377 | Gene names | S100G, CABP9K, CALB3, S100D | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-G (S100 calcium-binding protein G) (Vitamin D-dependent calcium-binding protein, intestinal) (CABP) (Calbindin D9K). | |||||
|
ONCO_HUMAN
|
||||||
| NC score | 0.079729 (rank : 39) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P32930 | Gene names | OCM, OCMN | |||
|
Domain Architecture |

|
|||||
| Description | Oncomodulin (OM) (Parvalbumin beta). | |||||
|
AIPL1_MOUSE
|
||||||
| NC score | 0.077831 (rank : 40) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q924K1 | Gene names | Aipl1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
AIPL1_HUMAN
|
||||||
| NC score | 0.076320 (rank : 41) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZN9, Q9H873, Q9NS10 | Gene names | AIPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl-hydrocarbon-interacting protein-like 1. | |||||
|
CABP3_HUMAN
|
||||||
| NC score | 0.072780 (rank : 42) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZU6, Q9NYY0, Q9NZT9 | Gene names | CABP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 3 (CaBP3). | |||||
|
OM34_MOUSE
|
||||||
| NC score | 0.072048 (rank : 43) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CYG7, Q3THD0, Q9D059 | Gene names | Tomm34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit). | |||||
|
PPID_MOUSE
|
||||||
| NC score | 0.071396 (rank : 44) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CR16, Q543G1 | Gene names | Ppid | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40). | |||||
|
OM34_HUMAN
|
||||||
| NC score | 0.071371 (rank : 45) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15785, Q9NTZ3 | Gene names | TOMM34 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial import receptor subunit TOM34 (Translocase of outer membrane 34 kDa subunit) (hTom34). | |||||
|
ONCO_MOUSE
|
||||||
| NC score | 0.071185 (rank : 46) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P51879, Q62004 | Gene names | Ocm | |||
|
Domain Architecture |

|
|||||
| Description | Oncomodulin (OM) (Parvalbumin beta). | |||||
|
RCN3_MOUSE
|
||||||
| NC score | 0.070804 (rank : 47) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BH97, Q58E50, Q8R137, Q99K35, Q9CTD4 | Gene names | Rcn3, D7Ertd671e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulocalbin-3 precursor. | |||||
|
PPID_HUMAN
|
||||||
| NC score | 0.069665 (rank : 48) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08752 | Gene names | PPID, CYP40, CYPD | |||
|
Domain Architecture |

|
|||||
| Description | 40 kDa peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-40) (CYP-40) (Cyclophilin-related protein). | |||||
|
RCN3_HUMAN
|
||||||
| NC score | 0.066951 (rank : 49) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96D15, Q9HBZ8 | Gene names | RCN3 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulocalbin-3 precursor (EF-hand calcium-binding protein RLP49). | |||||
|
TTC12_HUMAN
|
||||||
| NC score | 0.066914 (rank : 50) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
SPAG1_MOUSE
|
||||||
| NC score | 0.063576 (rank : 51) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80ZX8, Q8CCK7, Q9QZJ3, Q9QZJ4 | Gene names | Spag1, Tpis | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (TPR-containing protein involved in spermatogenesis) (TPIS). | |||||
|
SPAG1_HUMAN
|
||||||
| NC score | 0.060766 (rank : 52) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
STUB1_HUMAN
|
||||||
| NC score | 0.059549 (rank : 53) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
STUB1_MOUSE
|
||||||
| NC score | 0.057500 (rank : 54) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
CABP5_MOUSE
|
||||||
| NC score | 0.057150 (rank : 55) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
TTC12_MOUSE
|
||||||
| NC score | 0.056285 (rank : 56) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BW49 | Gene names | Ttc12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
SGTB_MOUSE
|
||||||
| NC score | 0.054758 (rank : 57) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VD33 | Gene names | Sgtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B. | |||||
|
TTC1_MOUSE
|
||||||
| NC score | 0.054484 (rank : 58) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
CABP1_HUMAN
|
||||||
| NC score | 0.052686 (rank : 59) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
CABP1_MOUSE
|
||||||
| NC score | 0.052670 (rank : 60) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
SGTB_HUMAN
|
||||||
| NC score | 0.052583 (rank : 61) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96EQ0 | Gene names | SGTB, SGT2 | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein B (Small glutamine-rich protein with tetratricopeptide repeats 2). | |||||
|
Z3H7B_HUMAN
|
||||||
| NC score | 0.051956 (rank : 62) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UGR2, Q5TFX9, Q8TBT9, Q9H8B6, Q9UGQ9, Q9UGR0, Q9UGR1, Q9UK03, Q9UPW9 | Gene names | ZC3H7B, KIAA1031 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7B (Rotavirus 'X'- associated non-structural protein) (RoXaN). | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.051205 (rank : 63) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
CABP5_HUMAN
|
||||||
| NC score | 0.051175 (rank : 64) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CALB2_HUMAN
|
||||||
| NC score | 0.040264 (rank : 65) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
CALB2_MOUSE
|
||||||
| NC score | 0.039839 (rank : 66) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
NCALD_HUMAN
|
||||||
| NC score | 0.038897 (rank : 67) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P61601, P29554, Q8IYC3, Q9H0W2 | Gene names | NCALD | |||
|
Domain Architecture |

|
|||||
| Description | Neurocalcin delta. | |||||
|
NCALD_MOUSE
|
||||||
| NC score | 0.038897 (rank : 68) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91X97, Q3TJS9, Q8BZN9 | Gene names | Ncald, D15Ertd412e | |||
|
Domain Architecture |

|
|||||
| Description | Neurocalcin delta. | |||||
|
HPCA_HUMAN
|
||||||
| NC score | 0.037205 (rank : 69) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P84074, P32076, P41211, P70510 | Gene names | HPCA, BDR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin (Calcium-binding protein BDR-2). | |||||
|
HPCA_MOUSE
|
||||||
| NC score | 0.037205 (rank : 70) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P84075, P32076, P41211, P70510 | Gene names | Hpca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin. | |||||
|
PLSI_HUMAN
|
||||||
| NC score | 0.033292 (rank : 71) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
HPCL1_MOUSE
|
||||||
| NC score | 0.032492 (rank : 72) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P62748, P35333 | Gene names | Hpcal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Neural visinin-like protein 3) (NVL-3) (NVP-3). | |||||
|
HPCL1_HUMAN
|
||||||
| NC score | 0.031847 (rank : 73) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P37235, Q969S5 | Gene names | HPCAL1, BDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Calcium-binding protein BDR-1) (HLP2). | |||||
|
DGKA_HUMAN
|
||||||
| NC score | 0.022945 (rank : 74) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23743, O75481, O75482, O75483 | Gene names | DGKA, DAGK, DAGK1 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase alpha (EC 2.7.1.107) (Diglyceride kinase alpha) (DGK-alpha) (DAG kinase alpha) (80 kDa diacylglycerol kinase). | |||||
|
S10A1_MOUSE
|
||||||
| NC score | 0.022893 (rank : 75) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56565, O88949 | Gene names | S100a1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein S100-A1 (S100 calcium-binding protein A1) (S-100 protein alpha subunit) (S-100 protein alpha chain). | |||||
|
NUCB2_MOUSE
|
||||||
| NC score | 0.010441 (rank : 76) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P81117 | Gene names | Nucb2, Nefa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA). | |||||
|
GLU2B_HUMAN
|
||||||
| NC score | 0.008775 (rank : 77) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
FSTL4_MOUSE
|
||||||
| NC score | 0.008339 (rank : 78) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
RBY1A_HUMAN
|
||||||
| NC score | 0.004769 (rank : 79) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||