Please be patient as the page loads
|
POLH_HUMAN
|
||||||
| SwissProt Accessions | Q9Y253, Q7L8E3, Q96BC4, Q9BX13 | Gene names | POLH, RAD30, RAD30A, XPV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase eta (EC 2.7.7.7) (RAD30 homolog A) (Xeroderma pigmentosum variant type protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
POLH_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y253, Q7L8E3, Q96BC4, Q9BX13 | Gene names | POLH, RAD30, RAD30A, XPV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase eta (EC 2.7.7.7) (RAD30 homolog A) (Xeroderma pigmentosum variant type protein). | |||||
|
POLH_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.984247 (rank : 2) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JJN0, Q9JJJ2 | Gene names | Polh, Rad30a, Xpv | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase eta (EC 2.7.7.7) (RAD30 homolog A) (Xeroderma pigmentosum variant type protein homolog). | |||||
|
POLI_HUMAN
|
||||||
| θ value | 1.9192e-34 (rank : 3) | NC score | 0.766083 (rank : 3) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UNA4, Q8N590, Q9H0S1, Q9NYH6 | Gene names | POLI, RAD30B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase iota (EC 2.7.7.7) (RAD30 homolog B) (Eta2). | |||||
|
POLI_MOUSE
|
||||||
| θ value | 2.77131e-33 (rank : 4) | NC score | 0.762270 (rank : 4) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6R3M4, Q641P1, Q6R3M3, Q9R1A6 | Gene names | Poli, Rad30b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase iota (EC 2.7.7.7) (Rad30 homolog B). | |||||
|
POLK_HUMAN
|
||||||
| θ value | 3.39556e-23 (rank : 5) | NC score | 0.672817 (rank : 6) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UBT6, Q86VJ8, Q8IZY0, Q8IZY1, Q8NB30, Q96L01, Q96Q86, Q96Q87, Q9UHC5 | Gene names | POLK, DINB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase kappa (EC 2.7.7.7) (DINB protein) (DINP). | |||||
|
POLK_MOUSE
|
||||||
| θ value | 5.79196e-23 (rank : 6) | NC score | 0.681310 (rank : 5) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QUG2, Q7TPY7 | Gene names | Polk, Dinb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase kappa (EC 2.7.7.7) (DINB protein) (DINP). | |||||
|
REV1_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 7) | NC score | 0.477741 (rank : 7) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBZ9, O95941, Q9C0J4, Q9NUP2 | Gene names | REV1L, REV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase) (Alpha integrin-binding protein 80) (AIBP80). | |||||
|
REV1_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 8) | NC score | 0.468484 (rank : 8) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q920Q2, Q9QXV2 | Gene names | Rev1l, Rev1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 9) | NC score | 0.059714 (rank : 9) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 10) | NC score | 0.057449 (rank : 10) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.011712 (rank : 39) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
EMSY_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.043344 (rank : 11) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.012764 (rank : 38) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MRIP_MOUSE
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.028470 (rank : 17) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
ORC1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.039454 (rank : 12) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1N2 | Gene names | Orc1l, Orc1 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 1. | |||||
|
DHI1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.032918 (rank : 15) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50172 | Gene names | Hsd11b1, Hsd11 | |||
|
Domain Architecture |
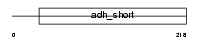
|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 1 (EC 1.1.1.146) (11-DH) (11-beta-hydroxysteroid dehydrogenase 1) (11-beta-HSD1) (11beta- HSD1A). | |||||
|
STAC_MOUSE
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.026541 (rank : 21) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97306 | Gene names | Stac | |||
|
Domain Architecture |
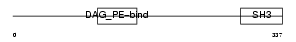
|
|||||
| Description | SH3 and cysteine-rich domain-containing protein (SRC homology 3 and cysteine-rich domain protein). | |||||
|
WNK3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.010251 (rank : 40) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.026809 (rank : 19) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SDCG1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.035026 (rank : 13) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CCP0, Q66JX6, Q8C9R6, Q8CA65, Q8JZT9, Q8R072, Q9CW30, Q9CYB8, Q9D4A9 | Gene names | Sdccag1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1. | |||||
|
STAT6_MOUSE
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.017178 (rank : 32) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P52633 | Gene names | Stat6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and transcription activator 6. | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.034006 (rank : 14) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
DHI1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.025998 (rank : 23) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28845 | Gene names | HSD11B1, HSD11, HSD11L | |||
|
Domain Architecture |
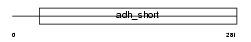
|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 1 (EC 1.1.1.146) (11-DH) (11-beta-hydroxysteroid dehydrogenase 1) (11-beta-HSD1). | |||||
|
PCIF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.025302 (rank : 24) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H4Z3, Q9NT85 | Gene names | PCIF1, C20orf67 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
PO121_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.027110 (rank : 18) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.009050 (rank : 42) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
DREB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.020337 (rank : 25) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QXS6, Q922X1, Q9QXS5 | Gene names | Dbn1, Drba | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
GP158_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.018622 (rank : 29) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.019778 (rank : 26) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.013590 (rank : 36) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
PATZ1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | -0.000093 (rank : 44) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 802 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBE1, Q9HBE2, Q9HBE3, Q9P1A9, Q9UDU0, Q9Y529 | Gene names | PATZ1, PATZ, RIAZ, ZBTB19, ZNF278, ZSG | |||
|
Domain Architecture |
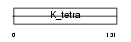
|
|||||
| Description | POZ-, AT hook-, and zinc finger-containing protein 1 (Zinc finger protein 278) (Zinc finger sarcoma gene protein) (BTB-POZ domain zinc finger transcription factor) (Protein kinase A RI-subunit alpha- associated protein) (Zinc finger and BTB domain-containing protein 19). | |||||
|
SPAG4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.014310 (rank : 34) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JJF2 | Gene names | Spag4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 4 protein (Outer dense fiber-associated protein SPAG4). | |||||
|
ALMS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.018755 (rank : 28) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
CEP57_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.014637 (rank : 33) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86XR8, Q14704, Q8IXP0, Q9BVF9 | Gene names | CEP57, KIAA0092, TSP57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin) (FGF2-interacting protein). | |||||
|
CIZ1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.018893 (rank : 27) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
CSF2R_MOUSE
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.018428 (rank : 31) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00941 | Gene names | Csf2ra, Csfgmra | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Granulocyte-macrophage colony-stimulating factor receptor alpha chain precursor (GM-CSF-R-alpha) (GMR) (CD116 antigen). | |||||
|
DDX58_HUMAN
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.026158 (rank : 22) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
MANBL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.026801 (rank : 20) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D8X0, Q3TDU6, Q91YV8 | Gene names | Manbal | |||
|
Domain Architecture |

|
|||||
| Description | Protein MANBAL. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.018461 (rank : 30) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SDCG1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.031768 (rank : 16) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60524, Q8WW70, Q9NWG1 | Gene names | SDCCAG1 | |||
|
Domain Architecture |
|
|||||
| Description | Serologically defined colon cancer antigen 1 (Antigen NY-CO-1). | |||||
|
CEP57_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.014293 (rank : 35) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
G6PD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.009697 (rank : 41) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11413, Q16000, Q16765, Q8IU70, Q8IU88, Q8IUA6, Q96PQ2 | Gene names | G6PD | |||
|
Domain Architecture |

|
|||||
| Description | Glucose-6-phosphate 1-dehydrogenase (EC 1.1.1.49) (G6PD). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.007545 (rank : 43) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
RAD18_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.013512 (rank : 37) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXK2, Q9CZB8 | Gene names | Rad18, Rad18sc | |||
|
Domain Architecture |

|
|||||
| Description | Postreplication repair protein RAD18 (mRAD18Sc). | |||||
|
POLH_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y253, Q7L8E3, Q96BC4, Q9BX13 | Gene names | POLH, RAD30, RAD30A, XPV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase eta (EC 2.7.7.7) (RAD30 homolog A) (Xeroderma pigmentosum variant type protein). | |||||
|
POLH_MOUSE
|
||||||
| NC score | 0.984247 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JJN0, Q9JJJ2 | Gene names | Polh, Rad30a, Xpv | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase eta (EC 2.7.7.7) (RAD30 homolog A) (Xeroderma pigmentosum variant type protein homolog). | |||||
|
POLI_HUMAN
|
||||||
| NC score | 0.766083 (rank : 3) | θ value | 1.9192e-34 (rank : 3) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UNA4, Q8N590, Q9H0S1, Q9NYH6 | Gene names | POLI, RAD30B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase iota (EC 2.7.7.7) (RAD30 homolog B) (Eta2). | |||||
|
POLI_MOUSE
|
||||||
| NC score | 0.762270 (rank : 4) | θ value | 2.77131e-33 (rank : 4) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6R3M4, Q641P1, Q6R3M3, Q9R1A6 | Gene names | Poli, Rad30b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase iota (EC 2.7.7.7) (Rad30 homolog B). | |||||
|
POLK_MOUSE
|
||||||
| NC score | 0.681310 (rank : 5) | θ value | 5.79196e-23 (rank : 6) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QUG2, Q7TPY7 | Gene names | Polk, Dinb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase kappa (EC 2.7.7.7) (DINB protein) (DINP). | |||||
|
POLK_HUMAN
|
||||||
| NC score | 0.672817 (rank : 6) | θ value | 3.39556e-23 (rank : 5) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UBT6, Q86VJ8, Q8IZY0, Q8IZY1, Q8NB30, Q96L01, Q96Q86, Q96Q87, Q9UHC5 | Gene names | POLK, DINB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase kappa (EC 2.7.7.7) (DINB protein) (DINP). | |||||
|
REV1_HUMAN
|
||||||
| NC score | 0.477741 (rank : 7) | θ value | 2.13673e-09 (rank : 7) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBZ9, O95941, Q9C0J4, Q9NUP2 | Gene names | REV1L, REV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase) (Alpha integrin-binding protein 80) (AIBP80). | |||||
|
REV1_MOUSE
|
||||||
| NC score | 0.468484 (rank : 8) | θ value | 2.79066e-09 (rank : 8) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q920Q2, Q9QXV2 | Gene names | Rev1l, Rev1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.059714 (rank : 9) | θ value | 0.0113563 (rank : 9) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.057449 (rank : 10) | θ value | 0.0193708 (rank : 10) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
EMSY_HUMAN
|
||||||
| NC score | 0.043344 (rank : 11) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
ORC1_MOUSE
|
||||||
| NC score | 0.039454 (rank : 12) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1N2 | Gene names | Orc1l, Orc1 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 1. | |||||
|
SDCG1_MOUSE
|
||||||
| NC score | 0.035026 (rank : 13) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CCP0, Q66JX6, Q8C9R6, Q8CA65, Q8JZT9, Q8R072, Q9CW30, Q9CYB8, Q9D4A9 | Gene names | Sdccag1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1. | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.034006 (rank : 14) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
DHI1_MOUSE
|
||||||
| NC score | 0.032918 (rank : 15) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50172 | Gene names | Hsd11b1, Hsd11 | |||
|
Domain Architecture |

|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 1 (EC 1.1.1.146) (11-DH) (11-beta-hydroxysteroid dehydrogenase 1) (11-beta-HSD1) (11beta- HSD1A). | |||||
|
SDCG1_HUMAN
|
||||||
| NC score | 0.031768 (rank : 16) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60524, Q8WW70, Q9NWG1 | Gene names | SDCCAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1 (Antigen NY-CO-1). | |||||
|
MRIP_MOUSE
|
||||||
| NC score | 0.028470 (rank : 17) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.027110 (rank : 18) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.026809 (rank : 19) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MANBL_MOUSE
|
||||||
| NC score | 0.026801 (rank : 20) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D8X0, Q3TDU6, Q91YV8 | Gene names | Manbal | |||
|
Domain Architecture |

|
|||||
| Description | Protein MANBAL. | |||||
|
STAC_MOUSE
|
||||||
| NC score | 0.026541 (rank : 21) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97306 | Gene names | Stac | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and cysteine-rich domain-containing protein (SRC homology 3 and cysteine-rich domain protein). | |||||
|
DDX58_HUMAN
|
||||||
| NC score | 0.026158 (rank : 22) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95786, Q5HYE1, Q5VYT1, Q9NT04 | Gene names | DDX58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
DHI1_HUMAN
|
||||||
| NC score | 0.025998 (rank : 23) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28845 | Gene names | HSD11B1, HSD11, HSD11L | |||
|
Domain Architecture |

|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 1 (EC 1.1.1.146) (11-DH) (11-beta-hydroxysteroid dehydrogenase 1) (11-beta-HSD1). | |||||
|
PCIF1_HUMAN
|
||||||
| NC score | 0.025302 (rank : 24) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H4Z3, Q9NT85 | Gene names | PCIF1, C20orf67 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylated CTD-interacting factor 1. | |||||
|
DREB_MOUSE
|
||||||
| NC score | 0.020337 (rank : 25) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QXS6, Q922X1, Q9QXS5 | Gene names | Dbn1, Drba | |||
|
Domain Architecture |

|
|||||
| Description | Drebrin (Developmentally-regulated brain protein). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.019778 (rank : 26) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CIZ1_HUMAN
|
||||||
| NC score | 0.018893 (rank : 27) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
ALMS1_HUMAN
|
||||||
| NC score | 0.018755 (rank : 28) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
GP158_HUMAN
|
||||||
| NC score | 0.018622 (rank : 29) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.018461 (rank : 30) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
CSF2R_MOUSE
|
||||||
| NC score | 0.018428 (rank : 31) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00941 | Gene names | Csf2ra, Csfgmra | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Granulocyte-macrophage colony-stimulating factor receptor alpha chain precursor (GM-CSF-R-alpha) (GMR) (CD116 antigen). | |||||
|
STAT6_MOUSE
|
||||||
| NC score | 0.017178 (rank : 32) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P52633 | Gene names | Stat6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and transcription activator 6. | |||||
|
CEP57_HUMAN
|
||||||
| NC score | 0.014637 (rank : 33) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86XR8, Q14704, Q8IXP0, Q9BVF9 | Gene names | CEP57, KIAA0092, TSP57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin) (FGF2-interacting protein). | |||||
|
SPAG4_MOUSE
|
||||||
| NC score | 0.014310 (rank : 34) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JJF2 | Gene names | Spag4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 4 protein (Outer dense fiber-associated protein SPAG4). | |||||
|
CEP57_MOUSE
|
||||||
| NC score | 0.014293 (rank : 35) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.013590 (rank : 36) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
RAD18_MOUSE
|
||||||
| NC score | 0.013512 (rank : 37) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXK2, Q9CZB8 | Gene names | Rad18, Rad18sc | |||
|
Domain Architecture |

|
|||||
| Description | Postreplication repair protein RAD18 (mRAD18Sc). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.012764 (rank : 38) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | 0.011712 (rank : 39) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
WNK3_HUMAN
|
||||||
| NC score | 0.010251 (rank : 40) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
G6PD_HUMAN
|
||||||
| NC score | 0.009697 (rank : 41) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11413, Q16000, Q16765, Q8IU70, Q8IU88, Q8IUA6, Q96PQ2 | Gene names | G6PD | |||
|
Domain Architecture |

|
|||||
| Description | Glucose-6-phosphate 1-dehydrogenase (EC 1.1.1.49) (G6PD). | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.009050 (rank : 42) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.007545 (rank : 43) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
PATZ1_HUMAN
|
||||||
| NC score | -0.000093 (rank : 44) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 44 | Target Neighborhood Hits | 802 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBE1, Q9HBE2, Q9HBE3, Q9P1A9, Q9UDU0, Q9Y529 | Gene names | PATZ1, PATZ, RIAZ, ZBTB19, ZNF278, ZSG | |||
|
Domain Architecture |

|
|||||
| Description | POZ-, AT hook-, and zinc finger-containing protein 1 (Zinc finger protein 278) (Zinc finger sarcoma gene protein) (BTB-POZ domain zinc finger transcription factor) (Protein kinase A RI-subunit alpha- associated protein) (Zinc finger and BTB domain-containing protein 19). | |||||