Please be patient as the page loads
|
NSDHL_HUMAN
|
||||||
| SwissProt Accessions | Q15738, O00344 | Gene names | NSDHL, H105E3 | |||
|
Domain Architecture |
|
|||||
| Description | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating (EC 1.1.1.170) (H105e3 protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NSDHL_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15738, O00344 | Gene names | NSDHL, H105E3 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating (EC 1.1.1.170) (H105e3 protein). | |||||
|
NSDHL_MOUSE
|
||||||
| θ value | 1.61255e-174 (rank : 2) | NC score | 0.997012 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R1J0, O55109 | Gene names | Nsdhl | |||
|
Domain Architecture |

|
|||||
| Description | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating (EC 1.1.1.170). | |||||
|
3BHS7_MOUSE
|
||||||
| θ value | 3.87602e-27 (rank : 3) | NC score | 0.838451 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9EQC1 | Gene names | Hsd3b7 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase type 7 (3 beta-hydroxysteroid dehydrogenase type VII) (3Beta-HSD VII) (3-beta-hydroxy-delta(5)-C27 steroid oxidoreductase) (EC 1.1.1.-) (C(27) 3beta-HSD). | |||||
|
3BHS6_MOUSE
|
||||||
| θ value | 5.06226e-27 (rank : 4) | NC score | 0.808017 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O35469 | Gene names | Hsd3b6 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase/delta 5-->4-isomerase type VI (3Beta-HSD VI) [Includes: 3-beta-hydroxy-delta(5)-steroid dehydrogenase (EC 1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase); Steroid delta-isomerase (EC 5.3.3.1) (Delta-5-3-ketosteroid isomerase)]. | |||||
|
3BHS3_MOUSE
|
||||||
| θ value | 6.61148e-27 (rank : 5) | NC score | 0.808824 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26150 | Gene names | Hsd3b3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase/delta 5-->4-isomerase type III (3Beta-HSD III) [Includes: 3-beta-hydroxy-delta(5)-steroid dehydrogenase (EC 1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase); Steroid delta-isomerase (EC 5.3.3.1) (Delta-5-3-ketosteroid isomerase)]. | |||||
|
3BHS2_MOUSE
|
||||||
| θ value | 2.51237e-26 (rank : 6) | NC score | 0.805198 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26149, Q8R0J6 | Gene names | Hsd3b2 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase/delta 5-->4-isomerase type II (3Beta-HSD II) [Includes: 3-beta-hydroxy-delta(5)-steroid dehydrogenase (EC 1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase); Steroid delta-isomerase (EC 5.3.3.1) (Delta-5-3-ketosteroid isomerase)]. | |||||
|
3BHS7_HUMAN
|
||||||
| θ value | 2.12685e-25 (rank : 7) | NC score | 0.832362 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H2F3, Q9BSN9 | Gene names | HSD3B7 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase type 7 (3 beta-hydroxysteroid dehydrogenase type VII) (3Beta-HSD VII) (3-beta-hydroxy-delta(5)-C27 steroid oxidoreductase) (EC 1.1.1.-) (C(27) 3beta-HSD). | |||||
|
3BHS1_MOUSE
|
||||||
| θ value | 6.84181e-24 (rank : 8) | NC score | 0.795951 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P24815, Q7TQ00 | Gene names | Hsd3b1, Hsd3b | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase/delta 5-->4-isomerase type I (3Beta-HSD I) [Includes: 3-beta-hydroxy-delta(5)-steroid dehydrogenase (EC 1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase); Steroid delta-isomerase (EC 5.3.3.1) (Delta- 5-3-ketosteroid isomerase)]. | |||||
|
3BHS2_HUMAN
|
||||||
| θ value | 3.39556e-23 (rank : 9) | NC score | 0.799984 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26439, Q16010, Q53GD4, Q6LDB9 | Gene names | HSD3B2, HSDB3B | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase/delta 5-->4-isomerase type II (3Beta-HSD II) [Includes: 3-beta-hydroxy-delta(5)-steroid dehydrogenase (EC 1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase); Steroid delta-isomerase (EC 5.3.3.1) (Delta-5-3-ketosteroid isomerase)]. | |||||
|
3BHS1_HUMAN
|
||||||
| θ value | 1.6852e-22 (rank : 10) | NC score | 0.797345 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P14060, Q14545, Q8IV65 | Gene names | HSD3B1, 3BH, HSDB3A | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase/delta 5-->4-isomerase type I (3Beta-HSD I) (Trophoblast antigen FDO161G) [Includes: 3-beta-hydroxy- delta(5)-steroid dehydrogenase (EC 1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase); Steroid delta- isomerase (EC 5.3.3.1) (Delta-5-3-ketosteroid isomerase)]. | |||||
|
3BHS5_MOUSE
|
||||||
| θ value | 1.74391e-19 (rank : 11) | NC score | 0.789096 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61694, Q91X27 | Gene names | Hsd3b5 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase type 5 (3 beta-hydroxysteroid dehydrogenase type V) (3Beta-HSD V) (NADPH-dependent 3-beta-hydroxy- delta(5)-steroid dehydrogenase) (EC 1.1.1.-) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase). | |||||
|
3BHS4_MOUSE
|
||||||
| θ value | 1.24977e-17 (rank : 12) | NC score | 0.780677 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61767 | Gene names | Hsd3b4 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase type 4 (3 beta-hydroxysteroid dehydrogenase type IV) (3Beta-HSD IV) (NADPH-dependent 3-beta-hydroxy- delta(5)-steroid dehydrogenase) (EC 1.1.1.-) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase). | |||||
|
UXS1_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 13) | NC score | 0.399742 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91XL3, Q3U854 | Gene names | Uxs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-glucuronic acid decarboxylase 1 (EC 4.1.1.35) (UDP-glucuronate decarboxylase 1) (UGD) (UXS-1). | |||||
|
UXS1_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 14) | NC score | 0.398869 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NBZ7, Q8NBX3, Q9H5C2 | Gene names | UXS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-glucuronic acid decarboxylase 1 (EC 4.1.1.35) (UDP-glucuronate decarboxylase 1) (UGD) (UXS-1). | |||||
|
TGDS_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 15) | NC score | 0.519026 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95455, Q5T3Z2, Q9H1T9 | Gene names | TGDS | |||
|
Domain Architecture |
|
|||||
| Description | dTDP-D-glucose 4,6-dehydratase (EC 4.2.1.46). | |||||
|
TGDS_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 16) | NC score | 0.518808 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VDR7 | Gene names | Tgds | |||
|
Domain Architecture |
|
|||||
| Description | dTDP-D-glucose 4,6-dehydratase (EC 4.2.1.46). | |||||
|
GALE_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 17) | NC score | 0.441911 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14376, Q86W41, Q9BVE3, Q9UJB4 | Gene names | GALE | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucose 4-epimerase (EC 5.1.3.2) (Galactowaldenase) (UDP-galactose 4-epimerase). | |||||
|
GALE_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 18) | NC score | 0.405262 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R059 | Gene names | Gale | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucose 4-epimerase (EC 5.1.3.2) (Galactowaldenase) (UDP-galactose 4-epimerase). | |||||
|
NDUA9_MOUSE
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.086093 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DC69, Q99JP9 | Gene names | Ndufa9 | |||
|
Domain Architecture |
|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 39 kDa subunit) (Complex I-39kD) (CI-39kD). | |||||
|
BLVRB_MOUSE
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.061545 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q923D2 | Gene names | Blvrb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin reductase (EC 1.5.1.30) (FR) (NADPH-dependent diaphorase) (NADPH-flavin reductase) (FLR) (Biliverdin reductase B) (EC 1.3.1.24) (BVR-B) (Biliverdin-IX beta-reductase). | |||||
|
NDUA9_HUMAN
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.066971 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16795, Q14076, Q2NKX0 | Gene names | NDUFA9, NDUFS2L | |||
|
Domain Architecture |
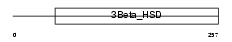
|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 39 kDa subunit) (Complex I-39kD) (CI-39kD). | |||||
|
IL16_MOUSE
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.016680 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54824, O70236, Q3TEM4, Q8R0G5 | Gene names | Il16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
AK1C1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.011075 (rank : 30) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04828, P52896, Q5SR15, Q9UCX2 | Gene names | AKR1C1, DDH, DDH1 | |||
|
Domain Architecture |
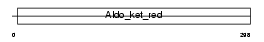
|
|||||
| Description | Aldo-keto reductase family 1 member C1 (EC 1.1.1.-) (20-alpha- hydroxysteroid dehydrogenase) (EC 1.1.1.149) (20-alpha-HSD) (Trans- 1,2-dihydrobenzene-1,2-diol dehydrogenase) (EC 1.3.1.20) (High- affinity hepatic bile acid-binding protein) (HBAB) (Chlordecone reductase homolog HAKRC) (Dihydrodiol dehydrogenase 1/2) (DD1/DD2). | |||||
|
AK1C4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.011155 (rank : 29) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17516, Q5T6A3, Q8WW84, Q9NS54 | Gene names | AKR1C4, CHDR | |||
|
Domain Architecture |

|
|||||
| Description | Aldo-keto reductase family 1 member C4 (EC 1.1.1.-) (Chlordecone reductase) (EC 1.1.1.225) (CDR) (3-alpha-hydroxysteroid dehydrogenase type I) (EC 1.1.1.50) (3-alpha-HSD1) (Dihydrodiol dehydrogenase 4) (DD4) (HAKRA). | |||||
|
AK1D1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.011205 (rank : 28) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51857 | Gene names | AKR1D1, SRD5B1 | |||
|
Domain Architecture |
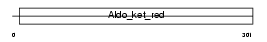
|
|||||
| Description | 3-oxo-5-beta-steroid 4-dehydrogenase (EC 1.3.1.3) (Delta(4)-3- ketosteroid 5-beta-reductase) (Aldo-keto reductase family 1 member D1). | |||||
|
AT12A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.002992 (rank : 31) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54707, Q13816, Q13817, Q16734 | Gene names | ATP12A, ATP1AL1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||
|
CRYL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.016794 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2S2, Q7Z4Z9 | Gene names | CRYL1, CRY | |||
|
Domain Architecture |
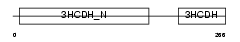
|
|||||
| Description | Lambda-crystallin homolog. | |||||
|
FCL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.097177 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13630, Q567Q9, Q9UDG7 | Gene names | TSTA3 | |||
|
Domain Architecture |
|
|||||
| Description | GDP-L-fucose synthetase (EC 1.1.1.271) (Protein FX) (Red cell NADP(H)- binding protein) (GDP-4-keto-6-deoxy-D-mannose-3,5-epimerase-4- reductase). | |||||
|
FCL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.090483 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23591 | Gene names | Tsta3, P35b, Tstap35b | |||
|
Domain Architecture |

|
|||||
| Description | GDP-L-fucose synthetase (EC 1.1.1.271) (Protein FX) (Red cell NADP(H)- binding protein) (GDP-4-keto-6-deoxy-D-mannose-3,5-epimerase-4- reductase) (Transplantation antigen P35B) (Tum-P35B antigen). | |||||
|
GMDS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.090453 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60547, O75357, Q5T954, Q6FH09, Q9UGZ3, Q9UJK9 | Gene names | GMDS | |||
|
Domain Architecture |
|
|||||
| Description | GDP-mannose 4,6 dehydratase (EC 4.2.1.47) (GDP-D-mannose dehydratase) (GMD). | |||||
|
GMDS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.088965 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K0C9 | Gene names | Gmds | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GDP-mannose 4,6 dehydratase (EC 4.2.1.47) (GDP-D-mannose dehydratase) (GMD). | |||||
|
NSDHL_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15738, O00344 | Gene names | NSDHL, H105E3 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating (EC 1.1.1.170) (H105e3 protein). | |||||
|
NSDHL_MOUSE
|
||||||
| NC score | 0.997012 (rank : 2) | θ value | 1.61255e-174 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R1J0, O55109 | Gene names | Nsdhl | |||
|
Domain Architecture |

|
|||||
| Description | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating (EC 1.1.1.170). | |||||
|
3BHS7_MOUSE
|
||||||
| NC score | 0.838451 (rank : 3) | θ value | 3.87602e-27 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9EQC1 | Gene names | Hsd3b7 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase type 7 (3 beta-hydroxysteroid dehydrogenase type VII) (3Beta-HSD VII) (3-beta-hydroxy-delta(5)-C27 steroid oxidoreductase) (EC 1.1.1.-) (C(27) 3beta-HSD). | |||||
|
3BHS7_HUMAN
|
||||||
| NC score | 0.832362 (rank : 4) | θ value | 2.12685e-25 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H2F3, Q9BSN9 | Gene names | HSD3B7 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase type 7 (3 beta-hydroxysteroid dehydrogenase type VII) (3Beta-HSD VII) (3-beta-hydroxy-delta(5)-C27 steroid oxidoreductase) (EC 1.1.1.-) (C(27) 3beta-HSD). | |||||
|
3BHS3_MOUSE
|
||||||
| NC score | 0.808824 (rank : 5) | θ value | 6.61148e-27 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26150 | Gene names | Hsd3b3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase/delta 5-->4-isomerase type III (3Beta-HSD III) [Includes: 3-beta-hydroxy-delta(5)-steroid dehydrogenase (EC 1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase); Steroid delta-isomerase (EC 5.3.3.1) (Delta-5-3-ketosteroid isomerase)]. | |||||
|
3BHS6_MOUSE
|
||||||
| NC score | 0.808017 (rank : 6) | θ value | 5.06226e-27 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O35469 | Gene names | Hsd3b6 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase/delta 5-->4-isomerase type VI (3Beta-HSD VI) [Includes: 3-beta-hydroxy-delta(5)-steroid dehydrogenase (EC 1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase); Steroid delta-isomerase (EC 5.3.3.1) (Delta-5-3-ketosteroid isomerase)]. | |||||
|
3BHS2_MOUSE
|
||||||
| NC score | 0.805198 (rank : 7) | θ value | 2.51237e-26 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26149, Q8R0J6 | Gene names | Hsd3b2 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase/delta 5-->4-isomerase type II (3Beta-HSD II) [Includes: 3-beta-hydroxy-delta(5)-steroid dehydrogenase (EC 1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase); Steroid delta-isomerase (EC 5.3.3.1) (Delta-5-3-ketosteroid isomerase)]. | |||||
|
3BHS2_HUMAN
|
||||||
| NC score | 0.799984 (rank : 8) | θ value | 3.39556e-23 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26439, Q16010, Q53GD4, Q6LDB9 | Gene names | HSD3B2, HSDB3B | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase/delta 5-->4-isomerase type II (3Beta-HSD II) [Includes: 3-beta-hydroxy-delta(5)-steroid dehydrogenase (EC 1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase); Steroid delta-isomerase (EC 5.3.3.1) (Delta-5-3-ketosteroid isomerase)]. | |||||
|
3BHS1_HUMAN
|
||||||
| NC score | 0.797345 (rank : 9) | θ value | 1.6852e-22 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P14060, Q14545, Q8IV65 | Gene names | HSD3B1, 3BH, HSDB3A | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase/delta 5-->4-isomerase type I (3Beta-HSD I) (Trophoblast antigen FDO161G) [Includes: 3-beta-hydroxy- delta(5)-steroid dehydrogenase (EC 1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase); Steroid delta- isomerase (EC 5.3.3.1) (Delta-5-3-ketosteroid isomerase)]. | |||||
|
3BHS1_MOUSE
|
||||||
| NC score | 0.795951 (rank : 10) | θ value | 6.84181e-24 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P24815, Q7TQ00 | Gene names | Hsd3b1, Hsd3b | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase/delta 5-->4-isomerase type I (3Beta-HSD I) [Includes: 3-beta-hydroxy-delta(5)-steroid dehydrogenase (EC 1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase); Steroid delta-isomerase (EC 5.3.3.1) (Delta- 5-3-ketosteroid isomerase)]. | |||||
|
3BHS5_MOUSE
|
||||||
| NC score | 0.789096 (rank : 11) | θ value | 1.74391e-19 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61694, Q91X27 | Gene names | Hsd3b5 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase type 5 (3 beta-hydroxysteroid dehydrogenase type V) (3Beta-HSD V) (NADPH-dependent 3-beta-hydroxy- delta(5)-steroid dehydrogenase) (EC 1.1.1.-) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase). | |||||
|
3BHS4_MOUSE
|
||||||
| NC score | 0.780677 (rank : 12) | θ value | 1.24977e-17 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61767 | Gene names | Hsd3b4 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase type 4 (3 beta-hydroxysteroid dehydrogenase type IV) (3Beta-HSD IV) (NADPH-dependent 3-beta-hydroxy- delta(5)-steroid dehydrogenase) (EC 1.1.1.-) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase). | |||||
|
TGDS_HUMAN
|
||||||
| NC score | 0.519026 (rank : 13) | θ value | 4.92598e-06 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95455, Q5T3Z2, Q9H1T9 | Gene names | TGDS | |||
|
Domain Architecture |

|
|||||
| Description | dTDP-D-glucose 4,6-dehydratase (EC 4.2.1.46). | |||||
|
TGDS_MOUSE
|
||||||
| NC score | 0.518808 (rank : 14) | θ value | 4.92598e-06 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VDR7 | Gene names | Tgds | |||
|
Domain Architecture |

|
|||||
| Description | dTDP-D-glucose 4,6-dehydratase (EC 4.2.1.46). | |||||
|
GALE_HUMAN
|
||||||
| NC score | 0.441911 (rank : 15) | θ value | 0.000158464 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14376, Q86W41, Q9BVE3, Q9UJB4 | Gene names | GALE | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucose 4-epimerase (EC 5.1.3.2) (Galactowaldenase) (UDP-galactose 4-epimerase). | |||||
|
GALE_MOUSE
|
||||||
| NC score | 0.405262 (rank : 16) | θ value | 0.00134147 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R059 | Gene names | Gale | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucose 4-epimerase (EC 5.1.3.2) (Galactowaldenase) (UDP-galactose 4-epimerase). | |||||
|
UXS1_MOUSE
|
||||||
| NC score | 0.399742 (rank : 17) | θ value | 2.69671e-12 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91XL3, Q3U854 | Gene names | Uxs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-glucuronic acid decarboxylase 1 (EC 4.1.1.35) (UDP-glucuronate decarboxylase 1) (UGD) (UXS-1). | |||||
|
UXS1_HUMAN
|
||||||
| NC score | 0.398869 (rank : 18) | θ value | 3.52202e-12 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NBZ7, Q8NBX3, Q9H5C2 | Gene names | UXS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-glucuronic acid decarboxylase 1 (EC 4.1.1.35) (UDP-glucuronate decarboxylase 1) (UGD) (UXS-1). | |||||
|
FCL_HUMAN
|
||||||
| NC score | 0.097177 (rank : 19) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13630, Q567Q9, Q9UDG7 | Gene names | TSTA3 | |||
|
Domain Architecture |

|
|||||
| Description | GDP-L-fucose synthetase (EC 1.1.1.271) (Protein FX) (Red cell NADP(H)- binding protein) (GDP-4-keto-6-deoxy-D-mannose-3,5-epimerase-4- reductase). | |||||
|
FCL_MOUSE
|
||||||
| NC score | 0.090483 (rank : 20) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23591 | Gene names | Tsta3, P35b, Tstap35b | |||
|
Domain Architecture |

|
|||||
| Description | GDP-L-fucose synthetase (EC 1.1.1.271) (Protein FX) (Red cell NADP(H)- binding protein) (GDP-4-keto-6-deoxy-D-mannose-3,5-epimerase-4- reductase) (Transplantation antigen P35B) (Tum-P35B antigen). | |||||
|
GMDS_HUMAN
|
||||||
| NC score | 0.090453 (rank : 21) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60547, O75357, Q5T954, Q6FH09, Q9UGZ3, Q9UJK9 | Gene names | GMDS | |||
|
Domain Architecture |

|
|||||
| Description | GDP-mannose 4,6 dehydratase (EC 4.2.1.47) (GDP-D-mannose dehydratase) (GMD). | |||||
|
GMDS_MOUSE
|
||||||
| NC score | 0.088965 (rank : 22) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K0C9 | Gene names | Gmds | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GDP-mannose 4,6 dehydratase (EC 4.2.1.47) (GDP-D-mannose dehydratase) (GMD). | |||||
|
NDUA9_MOUSE
|
||||||
| NC score | 0.086093 (rank : 23) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DC69, Q99JP9 | Gene names | Ndufa9 | |||
|
Domain Architecture |

|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 39 kDa subunit) (Complex I-39kD) (CI-39kD). | |||||
|
NDUA9_HUMAN
|
||||||
| NC score | 0.066971 (rank : 24) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16795, Q14076, Q2NKX0 | Gene names | NDUFA9, NDUFS2L | |||
|
Domain Architecture |

|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial precursor (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 39 kDa subunit) (Complex I-39kD) (CI-39kD). | |||||
|
BLVRB_MOUSE
|
||||||
| NC score | 0.061545 (rank : 25) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q923D2 | Gene names | Blvrb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin reductase (EC 1.5.1.30) (FR) (NADPH-dependent diaphorase) (NADPH-flavin reductase) (FLR) (Biliverdin reductase B) (EC 1.3.1.24) (BVR-B) (Biliverdin-IX beta-reductase). | |||||
|
CRYL1_HUMAN
|
||||||
| NC score | 0.016794 (rank : 26) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2S2, Q7Z4Z9 | Gene names | CRYL1, CRY | |||
|
Domain Architecture |

|
|||||
| Description | Lambda-crystallin homolog. | |||||
|
IL16_MOUSE
|
||||||
| NC score | 0.016680 (rank : 27) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54824, O70236, Q3TEM4, Q8R0G5 | Gene names | Il16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
AK1D1_HUMAN
|
||||||
| NC score | 0.011205 (rank : 28) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51857 | Gene names | AKR1D1, SRD5B1 | |||
|
Domain Architecture |
|
|||||
| Description | 3-oxo-5-beta-steroid 4-dehydrogenase (EC 1.3.1.3) (Delta(4)-3- ketosteroid 5-beta-reductase) (Aldo-keto reductase family 1 member D1). | |||||
|
AK1C4_HUMAN
|
||||||
| NC score | 0.011155 (rank : 29) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17516, Q5T6A3, Q8WW84, Q9NS54 | Gene names | AKR1C4, CHDR | |||
|
Domain Architecture |

|
|||||
| Description | Aldo-keto reductase family 1 member C4 (EC 1.1.1.-) (Chlordecone reductase) (EC 1.1.1.225) (CDR) (3-alpha-hydroxysteroid dehydrogenase type I) (EC 1.1.1.50) (3-alpha-HSD1) (Dihydrodiol dehydrogenase 4) (DD4) (HAKRA). | |||||
|
AK1C1_HUMAN
|
||||||
| NC score | 0.011075 (rank : 30) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04828, P52896, Q5SR15, Q9UCX2 | Gene names | AKR1C1, DDH, DDH1 | |||
|
Domain Architecture |
|
|||||
| Description | Aldo-keto reductase family 1 member C1 (EC 1.1.1.-) (20-alpha- hydroxysteroid dehydrogenase) (EC 1.1.1.149) (20-alpha-HSD) (Trans- 1,2-dihydrobenzene-1,2-diol dehydrogenase) (EC 1.3.1.20) (High- affinity hepatic bile acid-binding protein) (HBAB) (Chlordecone reductase homolog HAKRC) (Dihydrodiol dehydrogenase 1/2) (DD1/DD2). | |||||
|
AT12A_HUMAN
|
||||||
| NC score | 0.002992 (rank : 31) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54707, Q13816, Q13817, Q16734 | Gene names | ATP12A, ATP1AL1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium-transporting ATPase alpha chain 2 (EC 3.6.3.10) (Proton pump) (Non-gastric H(+)/K(+) ATPase subunit alpha). | |||||