Please be patient as the page loads
|
NGLY1_MOUSE
|
||||||
| SwissProt Accessions | Q9JI78, Q8K113, Q9CTK3 | Gene names | Ngly1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase (EC 3.5.1.52) (PNGase) (mPNGase) (Peptide:N-glycanase) (N-glycanase 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NGLY1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.990688 (rank : 2) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96IV0, Q59FB1, Q6PJD8, Q9BVR8, Q9NR70 | Gene names | NGLY1, PNG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase (EC 3.5.1.52) (PNGase) (hPNGase) (Peptide:N-glycanase) (N-glycanase 1). | |||||
|
NGLY1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JI78, Q8K113, Q9CTK3 | Gene names | Ngly1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase (EC 3.5.1.52) (PNGase) (mPNGase) (Peptide:N-glycanase) (N-glycanase 1). | |||||
|
UBXD1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 3) | NC score | 0.146154 (rank : 3) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZV1, Q96AH1, Q96IK9, Q9BZV0 | Gene names | UBXD1, UBXDC2 | |||
|
Domain Architecture |
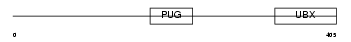
|
|||||
| Description | UBX domain-containing protein 1. | |||||
|
UBXD1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 4) | NC score | 0.135611 (rank : 4) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99PL6, Q91W79 | Gene names | Ubxd1, Ubxdc2 | |||
|
Domain Architecture |
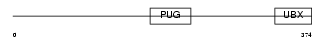
|
|||||
| Description | UBX domain-containing protein 1. | |||||
|
XPC_MOUSE
|
||||||
| θ value | 0.163984 (rank : 5) | NC score | 0.109258 (rank : 5) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51612, P54732, Q3TKI2, Q920M1, Q9DBW7 | Gene names | Xpc | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells homolog (Xeroderma pigmentosum group C-complementing protein homolog) (p125). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 0.279714 (rank : 6) | NC score | 0.026322 (rank : 9) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
XPC_HUMAN
|
||||||
| θ value | 0.279714 (rank : 7) | NC score | 0.106436 (rank : 6) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01831, Q96AX0 | Gene names | XPC, XPCC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells (Xeroderma pigmentosum group C-complementing protein) (p125). | |||||
|
ADIP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 8) | NC score | 0.059502 (rank : 7) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2D8, Q6P2P8, Q6ULS1, Q7L168, Q9UIX0 | Gene names | SSX2IP, KIAA0923 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Afadin- and alpha-actinin-binding protein (ADIP) (Afadin DIL domain- interacting protein) (SSX2-interacting protein). | |||||
|
DGKG_MOUSE
|
||||||
| θ value | 2.36792 (rank : 9) | NC score | 0.020945 (rank : 12) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WG7 | Gene names | Dgkg, Dagk3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma) (88 kDa diacylglycerol kinase). | |||||
|
ADIP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 10) | NC score | 0.057035 (rank : 8) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VC66, Q8BG59, Q8C7X0, Q8K2F7 | Gene names | Ssx2ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Afadin- and alpha-actinin-binding protein (ADIP) (Afadin DIL domain- interacting protein). | |||||
|
DGKG_HUMAN
|
||||||
| θ value | 4.03905 (rank : 11) | NC score | 0.019571 (rank : 13) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49619 | Gene names | DGKG, DAGK3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma). | |||||
|
GPNMB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 12) | NC score | 0.023644 (rank : 10) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14956, Q6UVX1, Q8N1A1 | Gene names | GPNMB, HGFIN, NMB | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane glycoprotein NMB precursor (Transmembrane glycoprotein HGFIN). | |||||
|
PLSB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 13) | NC score | 0.022682 (rank : 11) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61586 | Gene names | Gpam | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate acyltransferase, mitochondrial precursor (EC 2.3.1.15) (GPAT) (P90). | |||||
|
KRIT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 14) | NC score | 0.009852 (rank : 14) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00522, O43894, Q506L6, Q6U276, Q75N19, Q9H180, Q9H264, Q9HAX5 | Gene names | KRIT1, CCM1 | |||
|
Domain Architecture |
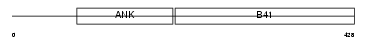
|
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein). | |||||
|
EMIL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 15) | NC score | 0.009112 (rank : 15) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K482 | Gene names | Emilin2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Basilin). | |||||
|
NGLY1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JI78, Q8K113, Q9CTK3 | Gene names | Ngly1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase (EC 3.5.1.52) (PNGase) (mPNGase) (Peptide:N-glycanase) (N-glycanase 1). | |||||
|
NGLY1_HUMAN
|
||||||
| NC score | 0.990688 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96IV0, Q59FB1, Q6PJD8, Q9BVR8, Q9NR70 | Gene names | NGLY1, PNG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase (EC 3.5.1.52) (PNGase) (hPNGase) (Peptide:N-glycanase) (N-glycanase 1). | |||||
|
UBXD1_HUMAN
|
||||||
| NC score | 0.146154 (rank : 3) | θ value | 0.0431538 (rank : 3) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZV1, Q96AH1, Q96IK9, Q9BZV0 | Gene names | UBXD1, UBXDC2 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 1. | |||||
|
UBXD1_MOUSE
|
||||||
| NC score | 0.135611 (rank : 4) | θ value | 0.125558 (rank : 4) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99PL6, Q91W79 | Gene names | Ubxd1, Ubxdc2 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 1. | |||||
|
XPC_MOUSE
|
||||||
| NC score | 0.109258 (rank : 5) | θ value | 0.163984 (rank : 5) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51612, P54732, Q3TKI2, Q920M1, Q9DBW7 | Gene names | Xpc | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells homolog (Xeroderma pigmentosum group C-complementing protein homolog) (p125). | |||||
|
XPC_HUMAN
|
||||||
| NC score | 0.106436 (rank : 6) | θ value | 0.279714 (rank : 7) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01831, Q96AX0 | Gene names | XPC, XPCC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells (Xeroderma pigmentosum group C-complementing protein) (p125). | |||||
|
ADIP_HUMAN
|
||||||
| NC score | 0.059502 (rank : 7) | θ value | 1.81305 (rank : 8) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2D8, Q6P2P8, Q6ULS1, Q7L168, Q9UIX0 | Gene names | SSX2IP, KIAA0923 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Afadin- and alpha-actinin-binding protein (ADIP) (Afadin DIL domain- interacting protein) (SSX2-interacting protein). | |||||
|
ADIP_MOUSE
|
||||||
| NC score | 0.057035 (rank : 8) | θ value | 3.0926 (rank : 10) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VC66, Q8BG59, Q8C7X0, Q8K2F7 | Gene names | Ssx2ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Afadin- and alpha-actinin-binding protein (ADIP) (Afadin DIL domain- interacting protein). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.026322 (rank : 9) | θ value | 0.279714 (rank : 6) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
GPNMB_HUMAN
|
||||||
| NC score | 0.023644 (rank : 10) | θ value | 4.03905 (rank : 12) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14956, Q6UVX1, Q8N1A1 | Gene names | GPNMB, HGFIN, NMB | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane glycoprotein NMB precursor (Transmembrane glycoprotein HGFIN). | |||||
|
PLSB_MOUSE
|
||||||
| NC score | 0.022682 (rank : 11) | θ value | 4.03905 (rank : 13) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61586 | Gene names | Gpam | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate acyltransferase, mitochondrial precursor (EC 2.3.1.15) (GPAT) (P90). | |||||
|
DGKG_MOUSE
|
||||||
| NC score | 0.020945 (rank : 12) | θ value | 2.36792 (rank : 9) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WG7 | Gene names | Dgkg, Dagk3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma) (88 kDa diacylglycerol kinase). | |||||
|
DGKG_HUMAN
|
||||||
| NC score | 0.019571 (rank : 13) | θ value | 4.03905 (rank : 11) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49619 | Gene names | DGKG, DAGK3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma). | |||||
|
KRIT1_HUMAN
|
||||||
| NC score | 0.009852 (rank : 14) | θ value | 6.88961 (rank : 14) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00522, O43894, Q506L6, Q6U276, Q75N19, Q9H180, Q9H264, Q9HAX5 | Gene names | KRIT1, CCM1 | |||
|
Domain Architecture |

|
|||||
| Description | Krev interaction trapped protein 1 (Krev interaction trapped 1) (Cerebral cavernous malformations 1 protein). | |||||
|
EMIL2_MOUSE
|
||||||
| NC score | 0.009112 (rank : 15) | θ value | 8.99809 (rank : 15) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K482 | Gene names | Emilin2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Basilin). | |||||