Please be patient as the page loads
|
MSPD2_MOUSE
|
||||||
| SwissProt Accessions | Q9CWP6, Q8BYF8, Q8BZB6, Q8C0G1, Q8R0T7 | Gene names | Mospd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Motile sperm domain-containing protein 2. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MSPD2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.986570 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8NHP6, Q8N3H2, Q8NA83 | Gene names | MOSPD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Motile sperm domain-containing protein 2. | |||||
|
MSPD2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9CWP6, Q8BYF8, Q8BZB6, Q8C0G1, Q8R0T7 | Gene names | Mospd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Motile sperm domain-containing protein 2. | |||||
|
VAPB_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 3) | NC score | 0.477903 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95292, O95293, Q9P0H0 | Gene names | VAPB | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein B/C (VAMP- associated protein B/C) (VAMP-B/VAMP-C) (VAP-B/VAP-C). | |||||
|
VAPB_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 4) | NC score | 0.460140 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QY76 | Gene names | Vapb | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein B (VAMP- associated protein B) (VAMP-associated protein 33b) (VAMP-B) (VAP-B). | |||||
|
VAPA_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 5) | NC score | 0.436318 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P0L0, O75453, Q5U0E7, Q9UBZ2 | Gene names | VAPA, VAP33 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein A (VAMP- associated protein A) (VAMP-A) (VAP-A) (33 kDa Vamp-associated protein) (VAP-33). | |||||
|
VAPA_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 6) | NC score | 0.431775 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WV55, Q3TJM1, Q9QY77 | Gene names | Vapa, Vap33 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein A (VAMP- associated protein A) (VAMP-A) (VAP-A) (33 kDa Vamp-associated protein) (VAP-33). | |||||
|
S14L4_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 7) | NC score | 0.166466 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R0F9 | Gene names | Sec14l4 | |||
|
Domain Architecture |
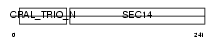
|
|||||
| Description | SEC14-like protein 4. | |||||
|
S14L4_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 8) | NC score | 0.155960 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UDX3 | Gene names | SEC14L4, TAP3 | |||
|
Domain Architecture |
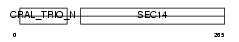
|
|||||
| Description | SEC14-like protein 4 (Tocopherol-associated protein 3). | |||||
|
PTN9_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 9) | NC score | 0.064696 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43378 | Gene names | PTPN9 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTN9_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 10) | NC score | 0.064744 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35239, Q7TSK0 | Gene names | Ptpn9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
MSPD1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 11) | NC score | 0.211348 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJG1, Q5H9C5, Q5H9C7 | Gene names | MOSPD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Motile sperm domain-containing protein 1. | |||||
|
MSPD1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 12) | NC score | 0.207049 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VEL0, Q497P5, Q9D8Y9 | Gene names | Mospd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Motile sperm domain-containing protein 1. | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 13) | NC score | 0.033204 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
MSPD3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 14) | NC score | 0.207476 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75425, O75423, O75424 | Gene names | MOSPD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Motile sperm domain-containing protein 3. | |||||
|
MSPD3_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 15) | NC score | 0.201633 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BGG6 | Gene names | Mospd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Motile sperm domain-containing protein 3. | |||||
|
S14L2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 16) | NC score | 0.113093 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O76054, Q9ULN4 | Gene names | SEC14L2, KIAA1186 | |||
|
Domain Architecture |
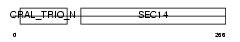
|
|||||
| Description | SEC14-like protein 2 (Alpha-tocopherol-associated protein) (TAP) (hTAP) (Supernatant protein factor) (SPF) (Squalene transfer protein). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 17) | NC score | 0.044272 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
E41L2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.022382 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
JHD1B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.031414 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.035878 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
GOGA5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.015381 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
SRPK2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.015063 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54781, Q8VCD9 | Gene names | Srpk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK2 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 2) (SR-protein-specific kinase 2) (SFRS protein kinase 2). | |||||
|
GLU2B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.038909 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
CD2L5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.003218 (rank : 56) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14004, Q6DKQ9, Q75MH4, Q75MH5, Q96JN4, Q9H4A0, Q9H4A1, Q9UDR4 | Gene names | CDC2L5, CDC2L, CHED, KIAA1791 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5) (Cholinesterase-related cell division controller). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.019015 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.021608 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
SPTA2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.010756 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.040623 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
DAXX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.033544 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.010262 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.030731 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.020954 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
KLF15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.002563 (rank : 57) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UIH9 | Gene names | KLF15, KKLF | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-like factor 15 (Kidney-enriched krueppel-like factor). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.047219 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.018998 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.006094 (rank : 55) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
PTN12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.023134 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05209, Q16130, Q59FD6, Q75MN8, Q86XU4 | Gene names | PTPN12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase G1) (PTPG1). | |||||
|
RBBP8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.021402 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99708, O75371 | Gene names | RBBP8, CTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 8 (RBBP-8) (CtBP-interacting protein) (CtIP) (Retinoblastoma-interacting protein and myosin-like) (RIM). | |||||
|
BRWD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.006514 (rank : 54) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
LIMA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.018996 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UHB6, Q2TAN7, Q9BVF2, Q9H8J1, Q9HBN5, Q9NX96, Q9NXC3, Q9NXU6, Q9P0H8, Q9UHB5 | Gene names | LIMA1, EPLIN, SREBP3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm). | |||||
|
XPP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.015288 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P1B1, Q91Y37 | Gene names | Xpnpep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Xaa-Pro aminopeptidase 1 (EC 3.4.11.9) (X-Pro aminopeptidase 1) (X- prolyl aminopeptidase 1, soluble) (Cytosolic aminopeptidase P) (Soluble aminopeptidase P) (sAmp) (Aminoacylproline aminopeptidase). | |||||
|
AVEN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.017017 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D9K3 | Gene names | Aven | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell death regulator Aven. | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.030066 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MPP10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.017611 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00566 | Gene names | MPHOSPH10, MPP10 | |||
|
Domain Architecture |

|
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
MYLK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.000229 (rank : 58) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H1R3, Q96I84 | Gene names | MYLK2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain kinase 2, skeletal/cardiac muscle (EC 2.7.11.18) (MLCK2). | |||||
|
PLTP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.011185 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55065, Q99L70 | Gene names | Pltp | |||
|
Domain Architecture |
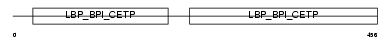
|
|||||
| Description | Phospholipid transfer protein precursor (Lipid transfer protein II). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.007253 (rank : 53) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PSIP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.017815 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
SGTA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.015095 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BJU0, Q8BGA6, Q99L52 | Gene names | Sgta, Sgt | |||
|
Domain Architecture |
|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A. | |||||
|
TEF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.010614 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLC6, Q3U426, Q3UGF4, Q3UM49, Q3URC8, Q6QHT6, Q8C6I0, Q8VD02 | Gene names | Tef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
CRAL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.051904 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P12271 | Gene names | RLBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cellular retinaldehyde-binding protein (CRALBP). | |||||
|
CT121_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.052427 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BTX7, Q5QPC1, Q9H1G2, Q9NQG8 | Gene names | C20orf121 | |||
|
Domain Architecture |
|
|||||
| Description | Protein C20orf121. | |||||
|
CT121_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.051481 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D3D0, Q3U0R3, Q7TS94 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Protein C20orf121 homolog. | |||||
|
S14L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.068040 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92503, Q99780 | Gene names | SEC14L1, SEC14L | |||
|
Domain Architecture |

|
|||||
| Description | SEC14-like protein 1. | |||||
|
S14L2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.089030 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99J08 | Gene names | Sec14l2 | |||
|
Domain Architecture |
|
|||||
| Description | SEC14-like protein 2 (Alpha-tocopherol-associated protein) (TAP). | |||||
|
S14L3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.094010 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UDX4 | Gene names | SEC14L3, TAP2 | |||
|
Domain Architecture |
|
|||||
| Description | SEC14-like protein 3 (Tocopherol-associated protein 2). | |||||
|
TTPA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.067619 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49638, Q71V64 | Gene names | TTPA, TPP1 | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-tocopherol transfer protein (Alpha-TTP). | |||||
|
TTPA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.064351 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BWP5, Q9CW51, Q9JL07 | Gene names | Ttpa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-tocopherol transfer protein (Alpha-TTP). | |||||
|
MSPD2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9CWP6, Q8BYF8, Q8BZB6, Q8C0G1, Q8R0T7 | Gene names | Mospd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Motile sperm domain-containing protein 2. | |||||
|
MSPD2_HUMAN
|
||||||
| NC score | 0.986570 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8NHP6, Q8N3H2, Q8NA83 | Gene names | MOSPD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Motile sperm domain-containing protein 2. | |||||
|
VAPB_HUMAN
|
||||||
| NC score | 0.477903 (rank : 3) | θ value | 5.08577e-11 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95292, O95293, Q9P0H0 | Gene names | VAPB | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein B/C (VAMP- associated protein B/C) (VAMP-B/VAMP-C) (VAP-B/VAP-C). | |||||
|
VAPB_MOUSE
|
||||||
| NC score | 0.460140 (rank : 4) | θ value | 2.13673e-09 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QY76 | Gene names | Vapb | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein B (VAMP- associated protein B) (VAMP-associated protein 33b) (VAMP-B) (VAP-B). | |||||
|
VAPA_HUMAN
|
||||||
| NC score | 0.436318 (rank : 5) | θ value | 2.61198e-07 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P0L0, O75453, Q5U0E7, Q9UBZ2 | Gene names | VAPA, VAP33 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein A (VAMP- associated protein A) (VAMP-A) (VAP-A) (33 kDa Vamp-associated protein) (VAP-33). | |||||
|
VAPA_MOUSE
|
||||||
| NC score | 0.431775 (rank : 6) | θ value | 4.45536e-07 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WV55, Q3TJM1, Q9QY77 | Gene names | Vapa, Vap33 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle-associated membrane protein-associated protein A (VAMP- associated protein A) (VAMP-A) (VAP-A) (33 kDa Vamp-associated protein) (VAP-33). | |||||
|
MSPD1_HUMAN
|
||||||
| NC score | 0.211348 (rank : 7) | θ value | 0.0193708 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJG1, Q5H9C5, Q5H9C7 | Gene names | MOSPD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Motile sperm domain-containing protein 1. | |||||
|
MSPD3_HUMAN
|
||||||
| NC score | 0.207476 (rank : 8) | θ value | 0.0431538 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75425, O75423, O75424 | Gene names | MOSPD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Motile sperm domain-containing protein 3. | |||||
|
MSPD1_MOUSE
|
||||||
| NC score | 0.207049 (rank : 9) | θ value | 0.0252991 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VEL0, Q497P5, Q9D8Y9 | Gene names | Mospd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Motile sperm domain-containing protein 1. | |||||
|
MSPD3_MOUSE
|
||||||
| NC score | 0.201633 (rank : 10) | θ value | 0.0563607 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BGG6 | Gene names | Mospd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Motile sperm domain-containing protein 3. | |||||
|
S14L4_MOUSE
|
||||||
| NC score | 0.166466 (rank : 11) | θ value | 0.00020696 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R0F9 | Gene names | Sec14l4 | |||
|
Domain Architecture |

|
|||||
| Description | SEC14-like protein 4. | |||||
|
S14L4_HUMAN
|
||||||
| NC score | 0.155960 (rank : 12) | θ value | 0.000602161 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UDX3 | Gene names | SEC14L4, TAP3 | |||
|
Domain Architecture |

|
|||||
| Description | SEC14-like protein 4 (Tocopherol-associated protein 3). | |||||
|
S14L2_HUMAN
|
||||||
| NC score | 0.113093 (rank : 13) | θ value | 0.0563607 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O76054, Q9ULN4 | Gene names | SEC14L2, KIAA1186 | |||
|
Domain Architecture |

|
|||||
| Description | SEC14-like protein 2 (Alpha-tocopherol-associated protein) (TAP) (hTAP) (Supernatant protein factor) (SPF) (Squalene transfer protein). | |||||
|
S14L3_HUMAN
|
||||||
| NC score | 0.094010 (rank : 14) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UDX4 | Gene names | SEC14L3, TAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SEC14-like protein 3 (Tocopherol-associated protein 2). | |||||
|
S14L2_MOUSE
|
||||||
| NC score | 0.089030 (rank : 15) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99J08 | Gene names | Sec14l2 | |||
|
Domain Architecture |

|
|||||
| Description | SEC14-like protein 2 (Alpha-tocopherol-associated protein) (TAP). | |||||
|
S14L1_HUMAN
|
||||||
| NC score | 0.068040 (rank : 16) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92503, Q99780 | Gene names | SEC14L1, SEC14L | |||
|
Domain Architecture |

|
|||||
| Description | SEC14-like protein 1. | |||||
|
TTPA_HUMAN
|
||||||
| NC score | 0.067619 (rank : 17) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49638, Q71V64 | Gene names | TTPA, TPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tocopherol transfer protein (Alpha-TTP). | |||||
|
PTN9_MOUSE
|
||||||
| NC score | 0.064744 (rank : 18) | θ value | 0.00509761 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35239, Q7TSK0 | Gene names | Ptpn9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
PTN9_HUMAN
|
||||||
| NC score | 0.064696 (rank : 19) | θ value | 0.00509761 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43378 | Gene names | PTPN9 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 9 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG2) (PTPase-MEG2). | |||||
|
TTPA_MOUSE
|
||||||
| NC score | 0.064351 (rank : 20) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BWP5, Q9CW51, Q9JL07 | Gene names | Ttpa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-tocopherol transfer protein (Alpha-TTP). | |||||
|
CT121_HUMAN
|
||||||
| NC score | 0.052427 (rank : 21) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BTX7, Q5QPC1, Q9H1G2, Q9NQG8 | Gene names | C20orf121 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C20orf121. | |||||
|
CRAL_HUMAN
|
||||||
| NC score | 0.051904 (rank : 22) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P12271 | Gene names | RLBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cellular retinaldehyde-binding protein (CRALBP). | |||||
|
CT121_MOUSE
|
||||||
| NC score | 0.051481 (rank : 23) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D3D0, Q3U0R3, Q7TS94 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C20orf121 homolog. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.047219 (rank : 24) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.044272 (rank : 25) | θ value | 0.0961366 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.040623 (rank : 26) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
GLU2B_HUMAN
|
||||||
| NC score | 0.038909 (rank : 27) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P14314, Q96BU9, Q96D06, Q9P0W9 | Gene names | PRKCSH, G19P1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.035878 (rank : 28) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
DAXX_HUMAN
|
||||||
| NC score | 0.033544 (rank : 29) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.033204 (rank : 30) | θ value | 0.0252991 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
JHD1B_MOUSE
|
||||||
| NC score | 0.031414 (rank : 31) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.030731 (rank : 32) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.030066 (rank : 33) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
PTN12_HUMAN
|
||||||
| NC score | 0.023134 (rank : 34) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05209, Q16130, Q59FD6, Q75MN8, Q86XU4 | Gene names | PTPN12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase G1) (PTPG1). | |||||
|
E41L2_HUMAN
|
||||||
| NC score | 0.022382 (rank : 35) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43491 | Gene names | EPB41L2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.021608 (rank : 36) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
RBBP8_HUMAN
|
||||||
| NC score | 0.021402 (rank : 37) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99708, O75371 | Gene names | RBBP8, CTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 8 (RBBP-8) (CtBP-interacting protein) (CtIP) (Retinoblastoma-interacting protein and myosin-like) (RIM). | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | 0.020954 (rank : 38) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.019015 (rank : 39) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.018998 (rank : 40) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
LIMA1_HUMAN
|
||||||
| NC score | 0.018996 (rank : 41) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UHB6, Q2TAN7, Q9BVF2, Q9H8J1, Q9HBN5, Q9NX96, Q9NXC3, Q9NXU6, Q9P0H8, Q9UHB5 | Gene names | LIMA1, EPLIN, SREBP3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm). | |||||
|
PSIP1_MOUSE
|
||||||
| NC score | 0.017815 (rank : 42) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99JF8, Q3TEJ7, Q3TTD7, Q3UTA1, Q80WQ7, Q99JF7, Q99LR4, Q9CT03 | Gene names | Psip1, Ledgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PC4 and SFRS1-interacting protein (Lens epithelium-derived growth factor) (mLEDGF). | |||||
|
MPP10_HUMAN
|
||||||
| NC score | 0.017611 (rank : 43) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00566 | Gene names | MPHOSPH10, MPP10 | |||
|
Domain Architecture |

|
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
AVEN_MOUSE
|
||||||
| NC score | 0.017017 (rank : 44) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D9K3 | Gene names | Aven | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell death regulator Aven. | |||||
|
GOGA5_HUMAN
|
||||||
| NC score | 0.015381 (rank : 45) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
XPP1_MOUSE
|
||||||
| NC score | 0.015288 (rank : 46) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P1B1, Q91Y37 | Gene names | Xpnpep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Xaa-Pro aminopeptidase 1 (EC 3.4.11.9) (X-Pro aminopeptidase 1) (X- prolyl aminopeptidase 1, soluble) (Cytosolic aminopeptidase P) (Soluble aminopeptidase P) (sAmp) (Aminoacylproline aminopeptidase). | |||||
|
SGTA_MOUSE
|
||||||
| NC score | 0.015095 (rank : 47) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BJU0, Q8BGA6, Q99L52 | Gene names | Sgta, Sgt | |||
|
Domain Architecture |

|
|||||
| Description | Small glutamine-rich tetratricopeptide repeat-containing protein A. | |||||
|
SRPK2_MOUSE
|
||||||
| NC score | 0.015063 (rank : 48) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54781, Q8VCD9 | Gene names | Srpk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK2 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 2) (SR-protein-specific kinase 2) (SFRS protein kinase 2). | |||||
|
PLTP_MOUSE
|
||||||
| NC score | 0.011185 (rank : 49) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55065, Q99L70 | Gene names | Pltp | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid transfer protein precursor (Lipid transfer protein II). | |||||
|
SPTA2_HUMAN
|
||||||
| NC score | 0.010756 (rank : 50) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
TEF_MOUSE
|
||||||
| NC score | 0.010614 (rank : 51) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLC6, Q3U426, Q3UGF4, Q3UM49, Q3URC8, Q6QHT6, Q8C6I0, Q8VD02 | Gene names | Tef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyrotroph embryonic factor. | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.010262 (rank : 52) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.007253 (rank : 53) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
BRWD1_MOUSE
|
||||||
| NC score | 0.006514 (rank : 54) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.006094 (rank : 55) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
CD2L5_HUMAN
|
||||||
| NC score | 0.003218 (rank : 56) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14004, Q6DKQ9, Q75MH4, Q75MH5, Q96JN4, Q9H4A0, Q9H4A1, Q9UDR4 | Gene names | CDC2L5, CDC2L, CHED, KIAA1791 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5) (Cholinesterase-related cell division controller). | |||||
|
KLF15_HUMAN
|
||||||
| NC score | 0.002563 (rank : 57) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UIH9 | Gene names | KLF15, KKLF | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-like factor 15 (Kidney-enriched krueppel-like factor). | |||||
|
MYLK2_HUMAN
|
||||||
| NC score | 0.000229 (rank : 58) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H1R3, Q96I84 | Gene names | MYLK2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain kinase 2, skeletal/cardiac muscle (EC 2.7.11.18) (MLCK2). | |||||