Please be patient as the page loads
|
MPIP1_HUMAN
|
||||||
| SwissProt Accessions | P30304, Q8IZH5, Q96IL3, Q9H2F2 | Gene names | CDC25A | |||
|
Domain Architecture |
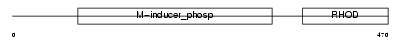
|
|||||
| Description | M-phase inducer phosphatase 1 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25A). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MPIP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30304, Q8IZH5, Q96IL3, Q9H2F2 | Gene names | CDC25A | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 1 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25A). | |||||
|
MPIP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.986521 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P48964 | Gene names | Cdc25a, Cdc25m3 | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 1 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25A). | |||||
|
MPIP2_MOUSE
|
||||||
| θ value | 9.01409e-109 (rank : 3) | NC score | 0.951804 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P30306, Q99LP3 | Gene names | Cdc25b, Cdc25m2 | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 2 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25B). | |||||
|
MPIP2_HUMAN
|
||||||
| θ value | 7.63087e-108 (rank : 4) | NC score | 0.957226 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P30305, O43551, Q13971, Q5JX77, Q6RSS1, Q9BRA6 | Gene names | CDC25B, CDC25HU2 | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 2 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25B). | |||||
|
MPIP3_HUMAN
|
||||||
| θ value | 7.71989e-68 (rank : 5) | NC score | 0.922894 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P30307, Q96PL3, Q9H168, Q9H2E8, Q9H2E9, Q9H2F1 | Gene names | CDC25C | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 3 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25C). | |||||
|
MPIP3_MOUSE
|
||||||
| θ value | 1.7238e-59 (rank : 6) | NC score | 0.916157 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48967, Q99KG6 | Gene names | Cdc25c, Cdc25m1 | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 3 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25C). | |||||
|
TLE2_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 7) | NC score | 0.058579 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04725, Q8WVY0, Q9Y6S0 | Gene names | TLE2 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 2 (ESG2). | |||||
|
TLE2_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 8) | NC score | 0.053896 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVB2 | Gene names | Tle2, Grg2 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 2. | |||||
|
CE110_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 9) | NC score | 0.055286 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TSH4, Q6A072 | Gene names | Cep110, Cp110, Kiaa0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
SFRS7_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 10) | NC score | 0.040366 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16629 | Gene names | SFRS7 | |||
|
Domain Architecture |
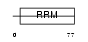
|
|||||
| Description | Splicing factor, arginine/serine-rich 7 (Splicing factor 9G8). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.011423 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
MISS_MOUSE
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.031597 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
SFRS7_MOUSE
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.029454 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BL97, Q8BMC6, Q8BUR2, Q8R2N4, Q8R3E9, Q91YS1 | Gene names | Sfrs7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 7. | |||||
|
UT14A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.019130 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BVJ6, Q5JYF1 | Gene names | UTP14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Antigen NY-CO- 16). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.015267 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
CHMP7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.030773 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WUX9, Q8NDM1, Q9BT50 | Gene names | CHMP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CHMP7. | |||||
|
CHMP7_MOUSE
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.031172 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R1T1, Q8CFW4 | Gene names | Chmp7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CHMP7. | |||||
|
DUS10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.020593 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6W6 | Gene names | DUSP10, MKP5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS10_MOUSE
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.020515 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ESS0, Q9CZY9 | Gene names | Dusp10, Mkp5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.007863 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
DACH2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.011973 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q925Q8, Q8BMA0 | Gene names | Dach2 | |||
|
Domain Architecture |
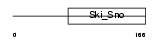
|
|||||
| Description | Dachshund homolog 2 (Dach2). | |||||
|
HAPIP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.006143 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60229 | Gene names | HAPIP, DUO | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein-interacting protein (Protein Duo). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.010126 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
ANPRB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.002432 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
ANPRB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.002703 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
ESCO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.007140 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
GP156_HUMAN
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.010115 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NFN8, Q86SN6 | Gene names | GPR156, GABABL, PGR28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor) (G-protein coupled receptor PGR28). | |||||
|
MPIP1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30304, Q8IZH5, Q96IL3, Q9H2F2 | Gene names | CDC25A | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 1 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25A). | |||||
|
MPIP1_MOUSE
|
||||||
| NC score | 0.986521 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P48964 | Gene names | Cdc25a, Cdc25m3 | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 1 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25A). | |||||
|
MPIP2_HUMAN
|
||||||
| NC score | 0.957226 (rank : 3) | θ value | 7.63087e-108 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P30305, O43551, Q13971, Q5JX77, Q6RSS1, Q9BRA6 | Gene names | CDC25B, CDC25HU2 | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 2 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25B). | |||||
|
MPIP2_MOUSE
|
||||||
| NC score | 0.951804 (rank : 4) | θ value | 9.01409e-109 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P30306, Q99LP3 | Gene names | Cdc25b, Cdc25m2 | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 2 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25B). | |||||
|
MPIP3_HUMAN
|
||||||
| NC score | 0.922894 (rank : 5) | θ value | 7.71989e-68 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P30307, Q96PL3, Q9H168, Q9H2E8, Q9H2E9, Q9H2F1 | Gene names | CDC25C | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 3 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25C). | |||||
|
MPIP3_MOUSE
|
||||||
| NC score | 0.916157 (rank : 6) | θ value | 1.7238e-59 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48967, Q99KG6 | Gene names | Cdc25c, Cdc25m1 | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 3 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25C). | |||||
|
TLE2_HUMAN
|
||||||
| NC score | 0.058579 (rank : 7) | θ value | 2.44474e-05 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04725, Q8WVY0, Q9Y6S0 | Gene names | TLE2 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 2 (ESG2). | |||||
|
CE110_MOUSE
|
||||||
| NC score | 0.055286 (rank : 8) | θ value | 0.0330416 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TSH4, Q6A072 | Gene names | Cep110, Cp110, Kiaa0419 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 110 kDa (Cep110 protein). | |||||
|
TLE2_MOUSE
|
||||||
| NC score | 0.053896 (rank : 9) | θ value | 0.00134147 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVB2 | Gene names | Tle2, Grg2 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 2. | |||||
|
SFRS7_HUMAN
|
||||||
| NC score | 0.040366 (rank : 10) | θ value | 0.0563607 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16629 | Gene names | SFRS7 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 7 (Splicing factor 9G8). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.031597 (rank : 11) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
CHMP7_MOUSE
|
||||||
| NC score | 0.031172 (rank : 12) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R1T1, Q8CFW4 | Gene names | Chmp7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CHMP7. | |||||
|
CHMP7_HUMAN
|
||||||
| NC score | 0.030773 (rank : 13) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WUX9, Q8NDM1, Q9BT50 | Gene names | CHMP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CHMP7. | |||||
|
SFRS7_MOUSE
|
||||||
| NC score | 0.029454 (rank : 14) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BL97, Q8BMC6, Q8BUR2, Q8R2N4, Q8R3E9, Q91YS1 | Gene names | Sfrs7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 7. | |||||
|
DUS10_HUMAN
|
||||||
| NC score | 0.020593 (rank : 15) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6W6 | Gene names | DUSP10, MKP5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS10_MOUSE
|
||||||
| NC score | 0.020515 (rank : 16) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ESS0, Q9CZY9 | Gene names | Dusp10, Mkp5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
UT14A_HUMAN
|
||||||
| NC score | 0.019130 (rank : 17) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BVJ6, Q5JYF1 | Gene names | UTP14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Antigen NY-CO- 16). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.015267 (rank : 18) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
DACH2_MOUSE
|
||||||
| NC score | 0.011973 (rank : 19) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q925Q8, Q8BMA0 | Gene names | Dach2 | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 2 (Dach2). | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.011423 (rank : 20) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.010126 (rank : 21) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
GP156_HUMAN
|
||||||
| NC score | 0.010115 (rank : 22) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NFN8, Q86SN6 | Gene names | GPR156, GABABL, PGR28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor) (G-protein coupled receptor PGR28). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.007863 (rank : 23) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
ESCO1_HUMAN
|
||||||
| NC score | 0.007140 (rank : 24) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
HAPIP_HUMAN
|
||||||
| NC score | 0.006143 (rank : 25) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60229 | Gene names | HAPIP, DUO | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein-interacting protein (Protein Duo). | |||||
|
ANPRB_MOUSE
|
||||||
| NC score | 0.002703 (rank : 26) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6VVW5, Q6VVW3, Q6VVW4, Q8CGA9 | Gene names | Npr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||
|
ANPRB_HUMAN
|
||||||
| NC score | 0.002432 (rank : 27) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20594, O60871, Q9UQ50 | Gene names | NPR2, ANPRB | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuretic peptide receptor B precursor (ANP-B) (ANPRB) (GC-B) (Guanylate cyclase B) (EC 4.6.1.2) (NPR-B) (Atrial natriuretic peptide B-type receptor). | |||||