Please be patient as the page loads
|
LN28B_HUMAN
|
||||||
| SwissProt Accessions | Q6ZN17 | Gene names | LIN28B, CSDD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog B. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LN28B_HUMAN
|
||||||
| θ value | 3.6343e-134 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q6ZN17 | Gene names | LIN28B, CSDD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog B. | |||||
|
LN28B_MOUSE
|
||||||
| θ value | 2.88631e-123 (rank : 2) | NC score | 0.985656 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q45KJ6, Q3UZC6, Q3V444 | Gene names | Lin28b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog B. | |||||
|
LN28A_MOUSE
|
||||||
| θ value | 3.4653e-68 (rank : 3) | NC score | 0.969224 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K3Y3, Q6NV62 | Gene names | Lin28, Lin28a, Tex17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog A (Testis-expressed protein 17). | |||||
|
LN28A_HUMAN
|
||||||
| θ value | 4.52582e-68 (rank : 4) | NC score | 0.969546 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H9Z2 | Gene names | LIN28, CSDD1, LIN28A, ZCCHC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog A (Zinc finger CCHC domain-containing protein 1). | |||||
|
YBOX2_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 5) | NC score | 0.626079 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2T7, Q8N4P0 | Gene names | YBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Y-box-binding protein 2 (Germ cell-specific Y-box-binding protein) (Contrin) (MSY2 homolog). | |||||
|
YBOX2_MOUSE
|
||||||
| θ value | 8.67504e-11 (rank : 6) | NC score | 0.621600 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z2C8, Q5NCW8, Q5NCW9, Q9Z2C7 | Gene names | Ybx2, Msy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Y-box-binding protein 2 (Germ cell-specific Y-box-binding protein) (FRGY2 homolog). | |||||
|
YBOX1_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 7) | NC score | 0.613202 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P67809, P16990, P16991, Q14972, Q15325, Q5FVF0 | Gene names | YBX1, NSEP1, YB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclease sensitive element-binding protein 1 (Y-box-binding protein 1) (Y-box transcription factor) (YB-1) (CCAAT-binding transcription factor I subunit A) (CBF-A) (Enhancer factor I subunit A) (EFI-A) (DNA-binding protein B) (DBPB). | |||||
|
YBOX1_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 8) | NC score | 0.615327 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P62960, P22568, P27817, P43482 | Gene names | Ybx1, Msy-1, Msy1, Nsep1, Yb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclease sensitive element-binding protein 1 (Y-box-binding protein 1) (Y-box transcription factor) (YB-1) (CCAAT-binding transcription factor I subunit A) (CBF-A) (Enhancer factor I subunit A) (EFI-A) (DNA-binding protein B) (DBPB). | |||||
|
DBPA_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 9) | NC score | 0.604302 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JKB3, Q80WG4, Q9EQF7, Q9EQF8 | Gene names | Csda, Msy4, Ybx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding protein A (Cold shock domain-containing protein A) (Y-box protein 3). | |||||
|
DBPA_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 10) | NC score | 0.598986 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P16989, Q14121, Q969N6 | Gene names | CSDA, DBPA | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein A (Cold shock domain-containing protein A) (Single-strand DNA-binding protein NF-GMB). | |||||
|
ZCH13_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 11) | NC score | 0.299906 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WW36 | Gene names | ZCCHC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 13. | |||||
|
CNBP_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 12) | NC score | 0.239552 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P62633, P20694, Q5QJR0, Q6PJI7, Q96NV3 | Gene names | CNBP, ZNF9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
CNBP_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 13) | NC score | 0.220436 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P53996, Q80Y06, Q8BP23 | Gene names | Cnbp, Cnbp1, Znf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
ZCH11_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 14) | NC score | 0.069806 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.023935 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
DDIT3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 16) | NC score | 0.064994 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35639, Q91YW9 | Gene names | Ddit3, Chop, Gadd153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
ZCHC7_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 17) | NC score | 0.227673 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N3Z6, Q5T0Q8, Q5T0Q9, Q5T0R0, Q8N2M1, Q8N4J2, Q8TBK8, Q9H648, Q9P0F0 | Gene names | ZCCHC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 7. | |||||
|
GAK3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.109418 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
TRPV3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.023901 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.040156 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
HXA11_MOUSE
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.013706 (rank : 69) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P31311, Q3V026 | Gene names | Hoxa11, Hox-1.9, Hoxa-11 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A11 (Hox-1.9). | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.106266 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.107526 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.106343 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK5_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.106054 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.024427 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.107131 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.105021 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK10_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.106108 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.105134 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
MARCS_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.050173 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
SFRS7_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.038535 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q16629 | Gene names | SFRS7 | |||
|
Domain Architecture |
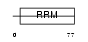
|
|||||
| Description | Splicing factor, arginine/serine-rich 7 (Splicing factor 9G8). | |||||
|
LOXL2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.029219 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y4K0, Q9BW70, Q9Y5Y8 | Gene names | LOXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2) (Lysyl oxidase-related protein 2) (Lysyl oxidase-related protein WS9-14). | |||||
|
LOXL2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.034370 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P58022, Q8BRE6, Q8BS86, Q8C4K0, Q9JJ39 | Gene names | Loxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2). | |||||
|
TENS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.034959 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.004333 (rank : 74) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
ZCHC8_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.082846 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
ZCHC9_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.160215 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8N567, Q9H027 | Gene names | ZCCHC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 9. | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.045059 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.045167 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
SFRS7_MOUSE
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.036444 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BL97, Q8BMC6, Q8BUR2, Q8R2N4, Q8R3E9, Q91YS1 | Gene names | Sfrs7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 7. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.037723 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TCF19_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.041582 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
ZCHC9_MOUSE
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.151007 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R1J3, Q921T6 | Gene names | Zcchc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 9. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.016532 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.030823 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
PGCB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.008887 (rank : 71) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.037561 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ZIC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.002248 (rank : 76) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95409, Q5VYA9, Q9H309 | Gene names | ZIC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZIC 2 (Zinc finger protein of the cerebellum 2). | |||||
|
ZIC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.002237 (rank : 77) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62520 | Gene names | Zic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZIC 2 (Zinc finger protein of the cerebellum 2). | |||||
|
HORN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.020220 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
SENP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.017689 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91ZX6, Q544T8, Q9D4Z0 | Gene names | Senp2, Smt3ip2 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 2 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP2) (SUMO-1/Smt3-specific isopeptidase 2) (Smt3ip2) (Axam2). | |||||
|
IF39_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.021816 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.026203 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.009561 (rank : 70) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
ZCHC8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.060525 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6NZY4, Q7L2P6, Q8N2K5, Q96SK7, Q9NSS2, Q9NSS3 | Gene names | ZCCHC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
JIP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.014851 (rank : 68) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ERE9, Q9CXI4 | Gene names | Mapk8ip2, Ib2, Jip2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.025028 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
PERQ1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.027830 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99MR1 | Gene names | Perq1 | |||
|
Domain Architecture |

|
|||||
| Description | PERQ amino acid rich with GYF domain protein 1. | |||||
|
CF061_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.028065 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NXL9, Q9HCV5 | Gene names | C6orf61 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf61. | |||||
|
CIZ1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.032674 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.017383 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
OS9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.020570 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13438, O00579 | Gene names | OS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein OS-9 precursor (Amplified in osteosarcoma 9). | |||||
|
SCUB3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.002380 (rank : 75) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IX30, Q5CZB3, Q86UZ9, Q8NAU9 | Gene names | SCUBE3, CEGF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.005298 (rank : 73) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
GAS7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.008528 (rank : 72) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60780, Q9QY25, Q9QY26, Q9QY34 | Gene names | Gas7 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
GAK13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.068929 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P62686 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1p13.3 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAK14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.071692 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P62687 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_16p3.3 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAK15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.077012 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P62688 | Gene names | ERVK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q21.2 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(I) Gag protein). | |||||
|
GAK17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.077418 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P62689 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein]. | |||||
|
GAK18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.085177 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK19_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.074238 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P62691 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_4p16.1 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAK7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.079338 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96PI4, Q9QC08 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K18 Gag protein) (HERV-K110 Gag protein) (HERV-K(C1a) Gag protein) [Contains: Matrix protein]. | |||||
|
GAK8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.088241 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.077304 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKH8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K104 Gag protein) [Contains: Matrix protein]. | |||||
|
IGEB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.051859 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P03975 | Gene names | Iap | |||
|
Domain Architecture |

|
|||||
| Description | IgE-binding protein. | |||||
|
ZCHC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.078647 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NUD5, Q6NT79 | Gene names | ZCCHC3, C20orf99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 3. | |||||
|
LN28B_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 3.6343e-134 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q6ZN17 | Gene names | LIN28B, CSDD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog B. | |||||
|
LN28B_MOUSE
|
||||||
| NC score | 0.985656 (rank : 2) | θ value | 2.88631e-123 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q45KJ6, Q3UZC6, Q3V444 | Gene names | Lin28b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog B. | |||||
|
LN28A_HUMAN
|
||||||
| NC score | 0.969546 (rank : 3) | θ value | 4.52582e-68 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H9Z2 | Gene names | LIN28, CSDD1, LIN28A, ZCCHC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog A (Zinc finger CCHC domain-containing protein 1). | |||||
|
LN28A_MOUSE
|
||||||
| NC score | 0.969224 (rank : 4) | θ value | 3.4653e-68 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8K3Y3, Q6NV62 | Gene names | Lin28, Lin28a, Tex17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-28 homolog A (Testis-expressed protein 17). | |||||
|
YBOX2_HUMAN
|
||||||
| NC score | 0.626079 (rank : 5) | θ value | 3.89403e-11 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2T7, Q8N4P0 | Gene names | YBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Y-box-binding protein 2 (Germ cell-specific Y-box-binding protein) (Contrin) (MSY2 homolog). | |||||
|
YBOX2_MOUSE
|
||||||
| NC score | 0.621600 (rank : 6) | θ value | 8.67504e-11 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z2C8, Q5NCW8, Q5NCW9, Q9Z2C7 | Gene names | Ybx2, Msy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Y-box-binding protein 2 (Germ cell-specific Y-box-binding protein) (FRGY2 homolog). | |||||
|
YBOX1_MOUSE
|
||||||
| NC score | 0.615327 (rank : 7) | θ value | 1.9326e-10 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P62960, P22568, P27817, P43482 | Gene names | Ybx1, Msy-1, Msy1, Nsep1, Yb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclease sensitive element-binding protein 1 (Y-box-binding protein 1) (Y-box transcription factor) (YB-1) (CCAAT-binding transcription factor I subunit A) (CBF-A) (Enhancer factor I subunit A) (EFI-A) (DNA-binding protein B) (DBPB). | |||||
|
YBOX1_HUMAN
|
||||||
| NC score | 0.613202 (rank : 8) | θ value | 1.9326e-10 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P67809, P16990, P16991, Q14972, Q15325, Q5FVF0 | Gene names | YBX1, NSEP1, YB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclease sensitive element-binding protein 1 (Y-box-binding protein 1) (Y-box transcription factor) (YB-1) (CCAAT-binding transcription factor I subunit A) (CBF-A) (Enhancer factor I subunit A) (EFI-A) (DNA-binding protein B) (DBPB). | |||||
|
DBPA_MOUSE
|
||||||
| NC score | 0.604302 (rank : 9) | θ value | 2.52405e-10 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JKB3, Q80WG4, Q9EQF7, Q9EQF8 | Gene names | Csda, Msy4, Ybx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding protein A (Cold shock domain-containing protein A) (Y-box protein 3). | |||||
|
DBPA_HUMAN
|
||||||
| NC score | 0.598986 (rank : 10) | θ value | 2.13673e-09 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P16989, Q14121, Q969N6 | Gene names | CSDA, DBPA | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein A (Cold shock domain-containing protein A) (Single-strand DNA-binding protein NF-GMB). | |||||
|
ZCH13_HUMAN
|
||||||
| NC score | 0.299906 (rank : 11) | θ value | 0.00228821 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WW36 | Gene names | ZCCHC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 13. | |||||
|
CNBP_HUMAN
|
||||||
| NC score | 0.239552 (rank : 12) | θ value | 0.0148317 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P62633, P20694, Q5QJR0, Q6PJI7, Q96NV3 | Gene names | CNBP, ZNF9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
ZCHC7_HUMAN
|
||||||
| NC score | 0.227673 (rank : 13) | θ value | 0.0961366 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N3Z6, Q5T0Q8, Q5T0Q9, Q5T0R0, Q8N2M1, Q8N4J2, Q8TBK8, Q9H648, Q9P0F0 | Gene names | ZCCHC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 7. | |||||
|
CNBP_MOUSE
|
||||||
| NC score | 0.220436 (rank : 14) | θ value | 0.0431538 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P53996, Q80Y06, Q8BP23 | Gene names | Cnbp, Cnbp1, Znf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
ZCHC9_HUMAN
|
||||||
| NC score | 0.160215 (rank : 15) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8N567, Q9H027 | Gene names | ZCCHC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 9. | |||||
|
ZCHC9_MOUSE
|
||||||
| NC score | 0.151007 (rank : 16) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R1J3, Q921T6 | Gene names | Zcchc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 9. | |||||
|
GAK3_HUMAN
|
||||||
| NC score | 0.109418 (rank : 17) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK1_HUMAN
|
||||||
| NC score | 0.107526 (rank : 18) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.107131 (rank : 19) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.106343 (rank : 20) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.106266 (rank : 21) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK10_HUMAN
|
||||||
| NC score | 0.106108 (rank : 22) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK5_HUMAN
|
||||||
| NC score | 0.106054 (rank : 23) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.105134 (rank : 24) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.105021 (rank : 25) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK8_HUMAN
|
||||||
| NC score | 0.088241 (rank : 26) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK18_HUMAN
|
||||||
| NC score | 0.085177 (rank : 27) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
ZCHC8_MOUSE
|
||||||
| NC score | 0.082846 (rank : 28) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
GAK7_HUMAN
|
||||||
| NC score | 0.079338 (rank : 29) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96PI4, Q9QC08 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K18 Gag protein) (HERV-K110 Gag protein) (HERV-K(C1a) Gag protein) [Contains: Matrix protein]. | |||||
|
ZCHC3_HUMAN
|
||||||
| NC score | 0.078647 (rank : 30) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NUD5, Q6NT79 | Gene names | ZCCHC3, C20orf99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 3. | |||||
|
GAK17_HUMAN
|
||||||
| NC score | 0.077418 (rank : 31) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P62689 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein]. | |||||
|
GAK9_HUMAN
|
||||||
| NC score | 0.077304 (rank : 32) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKH8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K104 Gag protein) [Contains: Matrix protein]. | |||||
|
GAK15_HUMAN
|
||||||
| NC score | 0.077012 (rank : 33) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P62688 | Gene names | ERVK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q21.2 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(I) Gag protein). | |||||
|
GAK19_HUMAN
|
||||||
| NC score | 0.074238 (rank : 34) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P62691 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_4p16.1 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAK14_HUMAN
|
||||||
| NC score | 0.071692 (rank : 35) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P62687 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_16p3.3 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
ZCH11_HUMAN
|
||||||
| NC score | 0.069806 (rank : 36) | θ value | 0.0563607 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
GAK13_HUMAN
|
||||||
| NC score | 0.068929 (rank : 37) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P62686 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1p13.3 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
DDIT3_MOUSE
|
||||||
| NC score | 0.064994 (rank : 38) | θ value | 0.0961366 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35639, Q91YW9 | Gene names | Ddit3, Chop, Gadd153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
ZCHC8_HUMAN
|
||||||
| NC score | 0.060525 (rank : 39) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6NZY4, Q7L2P6, Q8N2K5, Q96SK7, Q9NSS2, Q9NSS3 | Gene names | ZCCHC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
IGEB_MOUSE
|
||||||
| NC score | 0.051859 (rank : 40) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P03975 | Gene names | Iap | |||
|
Domain Architecture |

|
|||||
| Description | IgE-binding protein. | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.050173 (rank : 41) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.045167 (rank : 42) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.045059 (rank : 43) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
TCF19_HUMAN
|
||||||
| NC score | 0.041582 (rank : 44) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.040156 (rank : 45) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
SFRS7_HUMAN
|
||||||
| NC score | 0.038535 (rank : 46) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q16629 | Gene names | SFRS7 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 7 (Splicing factor 9G8). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.037723 (rank : 47) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.037561 (rank : 48) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SFRS7_MOUSE
|
||||||
| NC score | 0.036444 (rank : 49) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BL97, Q8BMC6, Q8BUR2, Q8R2N4, Q8R3E9, Q91YS1 | Gene names | Sfrs7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 7. | |||||
|
TENS1_HUMAN
|
||||||
| NC score | 0.034959 (rank : 50) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
LOXL2_MOUSE
|
||||||
| NC score | 0.034370 (rank : 51) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P58022, Q8BRE6, Q8BS86, Q8C4K0, Q9JJ39 | Gene names | Loxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2). | |||||
|
CIZ1_HUMAN
|
||||||
| NC score | 0.032674 (rank : 52) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.030823 (rank : 53) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
LOXL2_HUMAN
|
||||||
| NC score | 0.029219 (rank : 54) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y4K0, Q9BW70, Q9Y5Y8 | Gene names | LOXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2) (Lysyl oxidase-related protein 2) (Lysyl oxidase-related protein WS9-14). | |||||
|
CF061_HUMAN
|
||||||
| NC score | 0.028065 (rank : 55) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NXL9, Q9HCV5 | Gene names | C6orf61 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf61. | |||||
|
PERQ1_MOUSE
|
||||||
| NC score | 0.027830 (rank : 56) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99MR1 | Gene names | Perq1 | |||
|
Domain Architecture |

|
|||||
| Description | PERQ amino acid rich with GYF domain protein 1. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.026203 (rank : 57) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.025028 (rank : 58) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.024427 (rank : 59) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.023935 (rank : 60) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TRPV3_MOUSE
|
||||||
| NC score | 0.023901 (rank : 61) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
IF39_MOUSE
|
||||||
| NC score | 0.021816 (rank : 62) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
OS9_HUMAN
|
||||||
| NC score | 0.020570 (rank : 63) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13438, O00579 | Gene names | OS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein OS-9 precursor (Amplified in osteosarcoma 9). | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.020220 (rank : 64) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
SENP2_MOUSE
|
||||||
| NC score | 0.017689 (rank : 65) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91ZX6, Q544T8, Q9D4Z0 | Gene names | Senp2, Smt3ip2 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 2 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP2) (SUMO-1/Smt3-specific isopeptidase 2) (Smt3ip2) (Axam2). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.017383 (rank : 66) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.016532 (rank : 67) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
JIP2_MOUSE
|
||||||
| NC score | 0.014851 (rank : 68) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ERE9, Q9CXI4 | Gene names | Mapk8ip2, Ib2, Jip2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
HXA11_MOUSE
|
||||||
| NC score | 0.013706 (rank : 69) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P31311, Q3V026 | Gene names | Hoxa11, Hox-1.9, Hoxa-11 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A11 (Hox-1.9). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.009561 (rank : 70) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
PGCB_MOUSE
|
||||||
| NC score | 0.008887 (rank : 71) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
GAS7_MOUSE
|
||||||
| NC score | 0.008528 (rank : 72) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60780, Q9QY25, Q9QY26, Q9QY34 | Gene names | Gas7 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.005298 (rank : 73) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.004333 (rank : 74) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
SCUB3_HUMAN
|
||||||
| NC score | 0.002380 (rank : 75) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IX30, Q5CZB3, Q86UZ9, Q8NAU9 | Gene names | SCUBE3, CEGF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
ZIC2_HUMAN
|
||||||
| NC score | 0.002248 (rank : 76) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95409, Q5VYA9, Q9H309 | Gene names | ZIC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZIC 2 (Zinc finger protein of the cerebellum 2). | |||||
|
ZIC2_MOUSE
|
||||||
| NC score | 0.002237 (rank : 77) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62520 | Gene names | Zic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZIC 2 (Zinc finger protein of the cerebellum 2). | |||||