Please be patient as the page loads
|
GRHPR_HUMAN
|
||||||
| SwissProt Accessions | Q9UBQ7, Q9H3E9, Q9UKX1 | Gene names | GRHPR, GLXR | |||
|
Domain Architecture |
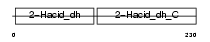
|
|||||
| Description | Glyoxylate reductase/hydroxypyruvate reductase (EC 1.1.1.79). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GRHPR_HUMAN
|
||||||
| θ value | 1.96665e-180 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UBQ7, Q9H3E9, Q9UKX1 | Gene names | GRHPR, GLXR | |||
|
Domain Architecture |

|
|||||
| Description | Glyoxylate reductase/hydroxypyruvate reductase (EC 1.1.1.79). | |||||
|
GRHPR_MOUSE
|
||||||
| θ value | 6.80647e-157 (rank : 2) | NC score | 0.998285 (rank : 2) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91Z53 | Gene names | Grhpr, Glxr | |||
|
Domain Architecture |
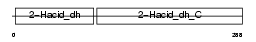
|
|||||
| Description | Glyoxylate reductase/hydroxypyruvate reductase (EC 1.1.1.79). | |||||
|
SERA_MOUSE
|
||||||
| θ value | 3.3877e-31 (rank : 3) | NC score | 0.818257 (rank : 4) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61753, Q75SV9 | Gene names | Phgdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-3-phosphoglycerate dehydrogenase (EC 1.1.1.95) (3-PGDH) (A10). | |||||
|
SERA_HUMAN
|
||||||
| θ value | 5.77852e-31 (rank : 4) | NC score | 0.818895 (rank : 3) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43175, Q5SZU3, Q9BQ01 | Gene names | PHGDH, PGDH3 | |||
|
Domain Architecture |
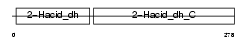
|
|||||
| Description | D-3-phosphoglycerate dehydrogenase (EC 1.1.1.95) (3-PGDH). | |||||
|
CTBP1_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 5) | NC score | 0.693983 (rank : 5) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13363 | Gene names | CTBP1, CTBP | |||
|
Domain Architecture |
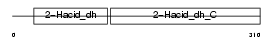
|
|||||
| Description | C-terminal-binding protein 1 (EC 1.1.1.-) (CtBP1). | |||||
|
CTBP1_MOUSE
|
||||||
| θ value | 1.80466e-16 (rank : 6) | NC score | 0.692874 (rank : 6) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88712, Q9QYG2 | Gene names | Ctbp1 | |||
|
Domain Architecture |
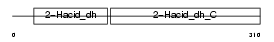
|
|||||
| Description | C-terminal-binding protein 1 (EC 1.1.1.-) (CtBP1). | |||||
|
CTBP2_MOUSE
|
||||||
| θ value | 5.25075e-16 (rank : 7) | NC score | 0.682192 (rank : 8) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P56546, O54855, O88462 | Gene names | Ctbp2 | |||
|
Domain Architecture |
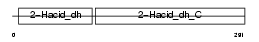
|
|||||
| Description | C-terminal-binding protein 2 (CtBP2). | |||||
|
CTBP2_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 8) | NC score | 0.682468 (rank : 7) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P56545, O43449 | Gene names | CTBP2 | |||
|
Domain Architecture |
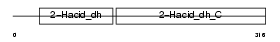
|
|||||
| Description | C-terminal-binding protein 2 (CtBP2). | |||||
|
3HIDH_MOUSE
|
||||||
| θ value | 2.36792 (rank : 9) | NC score | 0.031414 (rank : 9) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99L13, Q8BJY2 | Gene names | Hibadh | |||
|
Domain Architecture |
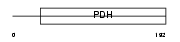
|
|||||
| Description | 3-hydroxyisobutyrate dehydrogenase, mitochondrial precursor (EC 1.1.1.31) (HIBADH). | |||||
|
SAHH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 10) | NC score | 0.023757 (rank : 11) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23526, Q96A36 | Gene names | AHCY, SAHH | |||
|
Domain Architecture |
|
|||||
| Description | Adenosylhomocysteinase (EC 3.3.1.1) (S-adenosyl-L-homocysteine hydrolase) (AdoHcyase). | |||||
|
JIP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 11) | NC score | 0.013740 (rank : 12) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q58A65, Q3UH77, Q3UHF0, Q58VQ4, Q5NC70, Q5NC78, Q6A057, Q6PAS3, Q8CJC2 | Gene names | Spag9, Jip4, Jsap2, Kiaa0516, Mapk8ip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (JNK/SAPK-associated protein 2) (JSAP2). | |||||
|
SAHH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 12) | NC score | 0.024974 (rank : 10) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50247, Q91WF1 | Gene names | Ahcy | |||
|
Domain Architecture |
|
|||||
| Description | Adenosylhomocysteinase (EC 3.3.1.1) (S-adenosyl-L-homocysteine hydrolase) (AdoHcyase) (Liver copper-binding protein) (CUBP). | |||||
|
JIP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 13) | NC score | 0.012791 (rank : 13) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60271, O60905, Q3KQU8, Q3MKM7, Q86WC7, Q86WC8, Q8IZX7, Q96II0, Q9H811 | Gene names | SPAG9, HSS, KIAA0516, MAPK8IP4, SYD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (Human lung cancer protein 6) (HLC-6) (Proliferation-inducing protein 6) (Sperm-specific protein) (Sperm surface protein) (Protein highly expressed in testis) (PHET) (Sunday driver 1). | |||||
|
GRHPR_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.96665e-180 (rank : 1) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UBQ7, Q9H3E9, Q9UKX1 | Gene names | GRHPR, GLXR | |||
|
Domain Architecture |

|
|||||
| Description | Glyoxylate reductase/hydroxypyruvate reductase (EC 1.1.1.79). | |||||
|
GRHPR_MOUSE
|
||||||
| NC score | 0.998285 (rank : 2) | θ value | 6.80647e-157 (rank : 2) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91Z53 | Gene names | Grhpr, Glxr | |||
|
Domain Architecture |

|
|||||
| Description | Glyoxylate reductase/hydroxypyruvate reductase (EC 1.1.1.79). | |||||
|
SERA_HUMAN
|
||||||
| NC score | 0.818895 (rank : 3) | θ value | 5.77852e-31 (rank : 4) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43175, Q5SZU3, Q9BQ01 | Gene names | PHGDH, PGDH3 | |||
|
Domain Architecture |

|
|||||
| Description | D-3-phosphoglycerate dehydrogenase (EC 1.1.1.95) (3-PGDH). | |||||
|
SERA_MOUSE
|
||||||
| NC score | 0.818257 (rank : 4) | θ value | 3.3877e-31 (rank : 3) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61753, Q75SV9 | Gene names | Phgdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-3-phosphoglycerate dehydrogenase (EC 1.1.1.95) (3-PGDH) (A10). | |||||
|
CTBP1_HUMAN
|
||||||
| NC score | 0.693983 (rank : 5) | θ value | 1.38178e-16 (rank : 5) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13363 | Gene names | CTBP1, CTBP | |||
|
Domain Architecture |

|
|||||
| Description | C-terminal-binding protein 1 (EC 1.1.1.-) (CtBP1). | |||||
|
CTBP1_MOUSE
|
||||||
| NC score | 0.692874 (rank : 6) | θ value | 1.80466e-16 (rank : 6) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88712, Q9QYG2 | Gene names | Ctbp1 | |||
|
Domain Architecture |

|
|||||
| Description | C-terminal-binding protein 1 (EC 1.1.1.-) (CtBP1). | |||||
|
CTBP2_HUMAN
|
||||||
| NC score | 0.682468 (rank : 7) | θ value | 6.85773e-16 (rank : 8) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P56545, O43449 | Gene names | CTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-terminal-binding protein 2 (CtBP2). | |||||
|
CTBP2_MOUSE
|
||||||
| NC score | 0.682192 (rank : 8) | θ value | 5.25075e-16 (rank : 7) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P56546, O54855, O88462 | Gene names | Ctbp2 | |||
|
Domain Architecture |

|
|||||
| Description | C-terminal-binding protein 2 (CtBP2). | |||||
|
3HIDH_MOUSE
|
||||||
| NC score | 0.031414 (rank : 9) | θ value | 2.36792 (rank : 9) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99L13, Q8BJY2 | Gene names | Hibadh | |||
|
Domain Architecture |

|
|||||
| Description | 3-hydroxyisobutyrate dehydrogenase, mitochondrial precursor (EC 1.1.1.31) (HIBADH). | |||||
|
SAHH_MOUSE
|
||||||
| NC score | 0.024974 (rank : 10) | θ value | 6.88961 (rank : 12) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50247, Q91WF1 | Gene names | Ahcy | |||
|
Domain Architecture |

|
|||||
| Description | Adenosylhomocysteinase (EC 3.3.1.1) (S-adenosyl-L-homocysteine hydrolase) (AdoHcyase) (Liver copper-binding protein) (CUBP). | |||||
|
SAHH_HUMAN
|
||||||
| NC score | 0.023757 (rank : 11) | θ value | 3.0926 (rank : 10) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23526, Q96A36 | Gene names | AHCY, SAHH | |||
|
Domain Architecture |

|
|||||
| Description | Adenosylhomocysteinase (EC 3.3.1.1) (S-adenosyl-L-homocysteine hydrolase) (AdoHcyase). | |||||
|
JIP4_MOUSE
|
||||||
| NC score | 0.013740 (rank : 12) | θ value | 6.88961 (rank : 11) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q58A65, Q3UH77, Q3UHF0, Q58VQ4, Q5NC70, Q5NC78, Q6A057, Q6PAS3, Q8CJC2 | Gene names | Spag9, Jip4, Jsap2, Kiaa0516, Mapk8ip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (JNK/SAPK-associated protein 2) (JSAP2). | |||||
|
JIP4_HUMAN
|
||||||
| NC score | 0.012791 (rank : 13) | θ value | 8.99809 (rank : 13) | |||
| Query Neighborhood Hits | 13 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60271, O60905, Q3KQU8, Q3MKM7, Q86WC7, Q86WC8, Q8IZX7, Q96II0, Q9H811 | Gene names | SPAG9, HSS, KIAA0516, MAPK8IP4, SYD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 4 (JNK-interacting protein 4) (JIP-4) (JNK-associated leucine-zipper protein) (JLP) (Sperm-associated antigen 9) (Mitogen-activated protein kinase 8- interacting protein 4) (Human lung cancer protein 6) (HLC-6) (Proliferation-inducing protein 6) (Sperm-specific protein) (Sperm surface protein) (Protein highly expressed in testis) (PHET) (Sunday driver 1). | |||||