Please be patient as the page loads
|
FBSH_MOUSE
|
||||||
| SwissProt Accessions | Q8R089 | Gene names | Fbs1, Fbs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable fibrosin-1 long transcript protein. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FBSH_MOUSE
|
||||||
| θ value | 1.33454e-136 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R089 | Gene names | Fbs1, Fbs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable fibrosin-1 long transcript protein. | |||||
|
FBSH_HUMAN
|
||||||
| θ value | 1.74701e-128 (rank : 2) | NC score | 0.985639 (rank : 2) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HAH7, Q96CI9, Q9H9X4 | Gene names | FBS1, FBS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable fibrosin-1 long transcript protein. | |||||
|
AUTS2_HUMAN
|
||||||
| θ value | 1.6247e-33 (rank : 3) | NC score | 0.642751 (rank : 3) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 4) | NC score | 0.096508 (rank : 8) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 5) | NC score | 0.062546 (rank : 29) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
INSM1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 6) | NC score | 0.037591 (rank : 65) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q63ZV0, Q9Z113 | Gene names | Insm1, Ia1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulinoma-associated protein 1 (Zinc finger protein IA-1). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.125558 (rank : 7) | NC score | 0.118857 (rank : 4) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.125558 (rank : 8) | NC score | 0.104169 (rank : 6) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CEBPE_HUMAN
|
||||||
| θ value | 0.279714 (rank : 9) | NC score | 0.069581 (rank : 17) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.086713 (rank : 9) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.029831 (rank : 71) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.047458 (rank : 61) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.066111 (rank : 22) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.020410 (rank : 82) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.067963 (rank : 20) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
PCD15_MOUSE
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.014115 (rank : 87) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PJ1 | Gene names | Pcdh15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
SON_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.047364 (rank : 62) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
EOMES_MOUSE
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.023983 (rank : 77) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54839, Q9QYG7 | Gene names | Eomes, Tbr2 | |||
|
Domain Architecture |
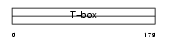
|
|||||
| Description | Eomesodermin homolog. | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.054722 (rank : 46) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
EOMES_HUMAN
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.023215 (rank : 80) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95936, Q8TAZ2, Q9UPM7 | Gene names | EOMES, TBR2 | |||
|
Domain Architecture |
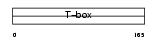
|
|||||
| Description | Eomesodermin homolog. | |||||
|
GASP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.032403 (rank : 68) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
HCFC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.053753 (rank : 49) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.106455 (rank : 5) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
NDF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.030559 (rank : 70) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.037924 (rank : 64) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PAK4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.006935 (rank : 92) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
RBM16_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.063140 (rank : 28) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
CN032_MOUSE
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.100493 (rank : 7) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
EP15R_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.020991 (rank : 81) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.043026 (rank : 63) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
NDF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.024565 (rank : 76) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.032232 (rank : 69) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
DSRAD_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.023346 (rank : 79) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99MU3, O70375, Q80UZ6, Q8C222, Q99MU2, Q99MU4, Q99MU7 | Gene names | Adar | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (RNA adenosine deaminase 1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.064974 (rank : 25) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
CTGE5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.012554 (rank : 91) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 928 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R311, Q8CIE3 | Gene names | Ctage5, Mea6, Mgea6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 homolog (cTAGE-5 protein) (Meningioma-expressed antigen 6). | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.018455 (rank : 84) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
PCGF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.023803 (rank : 78) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35227 | Gene names | PCGF2, MEL18, RNF110, ZNF144 | |||
|
Domain Architecture |
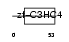
|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144). | |||||
|
WASF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.048963 (rank : 60) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R5H6, Q91W51, Q9ERQ9 | Gene names | Wasf1, Wave1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 1 (WASP-family protein member 1) (Protein WAVE-1). | |||||
|
BTBD7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.028063 (rank : 72) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CFE5, Q80TC3, Q8BZY9 | Gene names | Btbd7, Kiaa1525 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 7. | |||||
|
HPS6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.020201 (rank : 83) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BLY7, Q8BML5, Q8CIA3 | Gene names | Hps6, Ru | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 6 protein homolog (Ruby-eye protein) (Ru). | |||||
|
IASPP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.024653 (rank : 75) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.034871 (rank : 67) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
MAGD4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.014956 (rank : 86) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
MISS_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.071178 (rank : 15) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.035916 (rank : 66) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
SI1L3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.017339 (rank : 85) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
SNPC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.026692 (rank : 73) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13487, Q13486 | Gene names | SNAPC2, SNAP45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 2 (SNAPc subunit 2) (snRNA- activating protein complex 45 kDa subunit) (SNAPc 45 kDa subunit) (Small nuclear RNA-activating complex polypeptide 2) (Proximal sequence element-binding transcription factor subunit delta) (PSE- binding factor subunit delta) (PTF subunit delta). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.025265 (rank : 74) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
DCC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.013205 (rank : 90) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
FBX34_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.013443 (rank : 89) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NWN3, Q2VPB5, Q86TY4 | Gene names | FBXO34, FBX34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 34. | |||||
|
NOTC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.003530 (rank : 93) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
RBM25_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.014050 (rank : 88) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.069012 (rank : 19) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.050977 (rank : 57) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.061234 (rank : 30) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.075239 (rank : 10) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.055606 (rank : 41) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
ELN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.054852 (rank : 45) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15502, O15336, O15337, Q14233, Q14234, Q14235, Q14238, Q6P0L4, Q6ZWJ6, Q75MU5, Q7Z316, Q7Z3F5, Q9UMF5 | Gene names | ELN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
GGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.063781 (rank : 26) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GGN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.069860 (rank : 16) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.065691 (rank : 24) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.056771 (rank : 40) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.050288 (rank : 59) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.073303 (rank : 12) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.065759 (rank : 23) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.074617 (rank : 11) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PO121_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.054002 (rank : 48) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
PO121_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.051721 (rank : 54) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.055397 (rank : 42) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.054855 (rank : 44) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.063463 (rank : 27) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.073007 (rank : 13) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.072539 (rank : 14) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.066481 (rank : 21) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.052052 (rank : 52) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.051257 (rank : 56) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SELV_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.057154 (rank : 39) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.059924 (rank : 33) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.058993 (rank : 35) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SFR15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.050799 (rank : 58) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
SMR3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.059473 (rank : 34) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.053155 (rank : 50) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.058421 (rank : 36) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.057280 (rank : 38) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
T22D2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.059929 (rank : 32) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.069347 (rank : 18) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.055173 (rank : 43) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.052512 (rank : 51) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.051547 (rank : 55) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.057594 (rank : 37) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.054445 (rank : 47) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.051852 (rank : 53) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
ZN207_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.060162 (rank : 31) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
FBSH_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.33454e-136 (rank : 1) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R089 | Gene names | Fbs1, Fbs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable fibrosin-1 long transcript protein. | |||||
|
FBSH_HUMAN
|
||||||
| NC score | 0.985639 (rank : 2) | θ value | 1.74701e-128 (rank : 2) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9HAH7, Q96CI9, Q9H9X4 | Gene names | FBS1, FBS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable fibrosin-1 long transcript protein. | |||||
|
AUTS2_HUMAN
|
||||||
| NC score | 0.642751 (rank : 3) | θ value | 1.6247e-33 (rank : 3) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.118857 (rank : 4) | θ value | 0.125558 (rank : 7) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.106455 (rank : 5) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.104169 (rank : 6) | θ value | 0.125558 (rank : 8) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.100493 (rank : 7) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.096508 (rank : 8) | θ value | 6.43352e-06 (rank : 4) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.086713 (rank : 9) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.075239 (rank : 10) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.074617 (rank : 11) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.073303 (rank : 12) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.073007 (rank : 13) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.072539 (rank : 14) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.071178 (rank : 15) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.069860 (rank : 16) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
CEBPE_HUMAN
|
||||||
| NC score | 0.069581 (rank : 17) | θ value | 0.279714 (rank : 9) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.069347 (rank : 18) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.069012 (rank : 19) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.067963 (rank : 20) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.066481 (rank : 21) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.066111 (rank : 22) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.065759 (rank : 23) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.065691 (rank : 24) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.064974 (rank : 25) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.063781 (rank : 26) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.063463 (rank : 27) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RBM16_HUMAN
|
||||||
| NC score | 0.063140 (rank : 28) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.062546 (rank : 29) | θ value | 0.00102713 (rank : 5) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.061234 (rank : 30) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.060162 (rank : 31) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.059929 (rank : 32) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.059924 (rank : 33) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SMR3A_HUMAN
|
||||||
| NC score | 0.059473 (rank : 34) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.058993 (rank : 35) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.058421 (rank : 36) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.057594 (rank : 37) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.057280 (rank : 38) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SELV_HUMAN
|
||||||
| NC score | 0.057154 (rank : 39) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.056771 (rank : 40) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.055606 (rank : 41) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.055397 (rank : 42) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.055173 (rank : 43) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.054855 (rank : 44) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
ELN_HUMAN
|
||||||
| NC score | 0.054852 (rank : 45) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15502, O15336, O15337, Q14233, Q14234, Q14235, Q14238, Q6P0L4, Q6ZWJ6, Q75MU5, Q7Z316, Q7Z3F5, Q9UMF5 | Gene names | ELN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.054722 (rank : 46) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.054445 (rank : 47) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.054002 (rank : 48) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
HCFC1_HUMAN
|
||||||
| NC score | 0.053753 (rank : 49) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.053155 (rank : 50) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.052512 (rank : 51) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.052052 (rank : 52) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.051852 (rank : 53) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.051721 (rank : 54) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.051547 (rank : 55) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.051257 (rank : 56) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.050977 (rank : 57) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
SFR15_HUMAN
|
||||||
| NC score | 0.050799 (rank : 58) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.050288 (rank : 59) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
WASF1_MOUSE
|
||||||
| NC score | 0.048963 (rank : 60) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R5H6, Q91W51, Q9ERQ9 | Gene names | Wasf1, Wave1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 1 (WASP-family protein member 1) (Protein WAVE-1). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.047458 (rank : 61) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.047364 (rank : 62) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.043026 (rank : 63) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.037924 (rank : 64) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
INSM1_MOUSE
|
||||||
| NC score | 0.037591 (rank : 65) | θ value | 0.0961366 (rank : 6) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q63ZV0, Q9Z113 | Gene names | Insm1, Ia1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulinoma-associated protein 1 (Zinc finger protein IA-1). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.035916 (rank : 66) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.034871 (rank : 67) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
GASP2_MOUSE
|
||||||
| NC score | 0.032403 (rank : 68) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.032232 (rank : 69) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
NDF1_HUMAN
|
||||||
| NC score | 0.030559 (rank : 70) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.029831 (rank : 71) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
BTBD7_MOUSE
|
||||||
| NC score | 0.028063 (rank : 72) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CFE5, Q80TC3, Q8BZY9 | Gene names | Btbd7, Kiaa1525 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 7. | |||||
|
SNPC2_HUMAN
|
||||||
| NC score | 0.026692 (rank : 73) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13487, Q13486 | Gene names | SNAPC2, SNAP45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 2 (SNAPc subunit 2) (snRNA- activating protein complex 45 kDa subunit) (SNAPc 45 kDa subunit) (Small nuclear RNA-activating complex polypeptide 2) (Proximal sequence element-binding transcription factor subunit delta) (PSE- binding factor subunit delta) (PTF subunit delta). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.025265 (rank : 74) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
IASPP_HUMAN
|
||||||
| NC score | 0.024653 (rank : 75) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
NDF1_MOUSE
|
||||||
| NC score | 0.024565 (rank : 76) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
EOMES_MOUSE
|
||||||
| NC score | 0.023983 (rank : 77) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54839, Q9QYG7 | Gene names | Eomes, Tbr2 | |||
|
Domain Architecture |

|
|||||
| Description | Eomesodermin homolog. | |||||
|
PCGF2_HUMAN
|
||||||
| NC score | 0.023803 (rank : 78) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35227 | Gene names | PCGF2, MEL18, RNF110, ZNF144 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144). | |||||
|
DSRAD_MOUSE
|
||||||
| NC score | 0.023346 (rank : 79) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99MU3, O70375, Q80UZ6, Q8C222, Q99MU2, Q99MU4, Q99MU7 | Gene names | Adar | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (RNA adenosine deaminase 1). | |||||
|
EOMES_HUMAN
|
||||||
| NC score | 0.023215 (rank : 80) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95936, Q8TAZ2, Q9UPM7 | Gene names | EOMES, TBR2 | |||
|
Domain Architecture |

|
|||||
| Description | Eomesodermin homolog. | |||||
|
EP15R_HUMAN
|
||||||
| NC score | 0.020991 (rank : 81) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1083 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UBC2 | Gene names | EPS15L1, EPS15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.020410 (rank : 82) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
HPS6_MOUSE
|
||||||
| NC score | 0.020201 (rank : 83) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BLY7, Q8BML5, Q8CIA3 | Gene names | Hps6, Ru | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 6 protein homolog (Ruby-eye protein) (Ru). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.018455 (rank : 84) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
SI1L3_HUMAN
|
||||||
| NC score | 0.017339 (rank : 85) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
MAGD4_HUMAN
|
||||||
| NC score | 0.014956 (rank : 86) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
PCD15_MOUSE
|
||||||
| NC score | 0.014115 (rank : 87) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PJ1 | Gene names | Pcdh15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
RBM25_HUMAN
|
||||||
| NC score | 0.014050 (rank : 88) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
FBX34_HUMAN
|
||||||
| NC score | 0.013443 (rank : 89) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NWN3, Q2VPB5, Q86TY4 | Gene names | FBXO34, FBX34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 34. | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.013205 (rank : 90) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
CTGE5_MOUSE
|
||||||
| NC score | 0.012554 (rank : 91) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 928 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R311, Q8CIE3 | Gene names | Ctage5, Mea6, Mgea6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 homolog (cTAGE-5 protein) (Meningioma-expressed antigen 6). | |||||
|
PAK4_MOUSE
|
||||||
| NC score | 0.006935 (rank : 92) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
NOTC2_MOUSE
|
||||||
| NC score | 0.003530 (rank : 93) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 52 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||