Please be patient as the page loads
|
FACE1_MOUSE
|
||||||
| SwissProt Accessions | Q80W54, Q8BJK4, Q8K569 | Gene names | Zmpste24, Face1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAAX prenyl protease 1 homolog (EC 3.4.24.84) (Prenyl protein-specific endoprotease 1) (Farnesylated proteins-converting enzyme 1) (FACE-1) (Zinc metalloproteinase Ste24 homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FACE1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997361 (rank : 2) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75844, Q8NDZ8, Q9UBQ2 | Gene names | ZMPSTE24, FACE1, STE24 | |||
|
Domain Architecture |
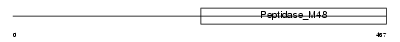
|
|||||
| Description | CAAX prenyl protease 1 homolog (EC 3.4.24.84) (Prenyl protein-specific endoprotease 1) (Farnesylated proteins-converting enzyme 1) (FACE-1) (Zinc metalloproteinase Ste24 homolog). | |||||
|
FACE1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80W54, Q8BJK4, Q8K569 | Gene names | Zmpste24, Face1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAAX prenyl protease 1 homolog (EC 3.4.24.84) (Prenyl protein-specific endoprotease 1) (Farnesylated proteins-converting enzyme 1) (FACE-1) (Zinc metalloproteinase Ste24 homolog). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 3) | NC score | 0.027496 (rank : 3) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 4) | NC score | 0.012177 (rank : 6) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 5) | NC score | 0.005359 (rank : 8) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
MYPT2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 6) | NC score | 0.007565 (rank : 7) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
PSPB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 7) | NC score | 0.020501 (rank : 4) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07988, Q96R04 | Gene names | SFTPB, SFTP3 | |||
|
Domain Architecture |
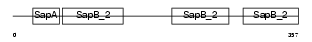
|
|||||
| Description | Pulmonary surfactant-associated protein B precursor (SP-B) (6 kDa protein) (Pulmonary surfactant-associated proteolipid SPL(Phe)) (18 kDa pulmonary-surfactant protein). | |||||
|
TGM3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 8) | NC score | 0.013250 (rank : 5) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08188, O95933 | Gene names | TGM3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase E precursor (EC 2.3.2.13) (TGase E) (TGE) (TG(E)) (Transglutaminase-3) [Contains: Protein- glutamine gamma-glutamyltransferase E 50 kDa non-catalytic chain; Protein-glutamine gamma-glutamyltransferase E 27 kDa catalytic chain]. | |||||
|
FACE1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80W54, Q8BJK4, Q8K569 | Gene names | Zmpste24, Face1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAAX prenyl protease 1 homolog (EC 3.4.24.84) (Prenyl protein-specific endoprotease 1) (Farnesylated proteins-converting enzyme 1) (FACE-1) (Zinc metalloproteinase Ste24 homolog). | |||||
|
FACE1_HUMAN
|
||||||
| NC score | 0.997361 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75844, Q8NDZ8, Q9UBQ2 | Gene names | ZMPSTE24, FACE1, STE24 | |||
|
Domain Architecture |

|
|||||
| Description | CAAX prenyl protease 1 homolog (EC 3.4.24.84) (Prenyl protein-specific endoprotease 1) (Farnesylated proteins-converting enzyme 1) (FACE-1) (Zinc metalloproteinase Ste24 homolog). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.027496 (rank : 3) | θ value | 0.813845 (rank : 3) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
PSPB_HUMAN
|
||||||
| NC score | 0.020501 (rank : 4) | θ value | 6.88961 (rank : 7) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07988, Q96R04 | Gene names | SFTPB, SFTP3 | |||
|
Domain Architecture |

|
|||||
| Description | Pulmonary surfactant-associated protein B precursor (SP-B) (6 kDa protein) (Pulmonary surfactant-associated proteolipid SPL(Phe)) (18 kDa pulmonary-surfactant protein). | |||||
|
TGM3_HUMAN
|
||||||
| NC score | 0.013250 (rank : 5) | θ value | 6.88961 (rank : 8) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08188, O95933 | Gene names | TGM3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-glutamine gamma-glutamyltransferase E precursor (EC 2.3.2.13) (TGase E) (TGE) (TG(E)) (Transglutaminase-3) [Contains: Protein- glutamine gamma-glutamyltransferase E 50 kDa non-catalytic chain; Protein-glutamine gamma-glutamyltransferase E 27 kDa catalytic chain]. | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.012177 (rank : 6) | θ value | 2.36792 (rank : 4) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
MYPT2_HUMAN
|
||||||
| NC score | 0.007565 (rank : 7) | θ value | 5.27518 (rank : 6) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.005359 (rank : 8) | θ value | 3.0926 (rank : 5) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||