Please be patient as the page loads
|
COE4_HUMAN
|
||||||
| SwissProt Accessions | Q9BQW3, Q5JY53, Q9NUB6, Q9P2A6 | Gene names | EBF4, COE4, KIAA1442 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE4 (Early B-cell factor 4) (EBF-4) (Olf-1/EBF- like 4) (OE-4) (O/E-4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
COE1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993607 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UH73, Q8IW11 | Gene names | EBF1, COE1, EBF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
COE1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993607 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07802 | Gene names | Ebf1, Coe1, Ebf | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
COE2_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.994740 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08792, Q543D5 | Gene names | Ebf2, Coe2, Mmot1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE2 (Early B-cell factor 2) (EBF-2) (Olf-1/EBF- like 3) (OE-3) (O/E-3) (Metencephalon-mesencephalon-olfactory transcription factor 1) (MET-mesencephalon-olfactory transcription factor 1) (MET-mesencephalon-olfactory TF1). | |||||
|
COE3_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.991072 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H4W6, Q5T6H9, Q9H4W5 | Gene names | EBF3, COE3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
COE3_MOUSE
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.991308 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08791, O08793 | Gene names | Ebf3, Coe3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
COE4_HUMAN
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BQW3, Q5JY53, Q9NUB6, Q9P2A6 | Gene names | EBF4, COE4, KIAA1442 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE4 (Early B-cell factor 4) (EBF-4) (Olf-1/EBF- like 4) (OE-4) (O/E-4). | |||||
|
COE4_MOUSE
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.997630 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K4J2, Q8K4J1, Q8K4J3, Q8K4J4, Q8K4J5 | Gene names | Ebf4, Coe4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE4 (Early B-cell factor 4) (EBF-4) (Olf-1/EBF- like 4) (OE-4) (O/E-4). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 8) | NC score | 0.046086 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 9) | NC score | 0.013347 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
UBQL3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 10) | NC score | 0.018338 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H347, Q9NRE0 | Gene names | UBQLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 1.38821 (rank : 11) | NC score | 0.015390 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
GLI1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 12) | NC score | 0.001686 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||
|
CCNL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.019143 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96S94, Q5T2N5, Q5T2N6, Q6IQ12, Q7Z4Z8, Q8N3C9, Q8N3D5, Q8NHE3, Q8TEL0, Q96B00 | Gene names | CCNL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Paneth cell-enhanced expression protein). | |||||
|
ASTL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.010262 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
IRK10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 15) | NC score | 0.019543 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78508, Q8N4I7, Q92808 | Gene names | KCNJ10 | |||
|
Domain Architecture |
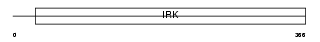
|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 10 (Potassium channel, inwardly rectifying subfamily J member 10) (Inward rectifier K(+) channel Kir1.2) (ATP-dependent inwardly rectifying potassium channel Kir4.1). | |||||
|
PIGQ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.020555 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BRB3, O14927, Q96G00, Q96S22, Q9UJH4 | Gene names | PIGQ, GPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q (EC 2.4.1.198) (Phosphatidylinositol-glycan biosynthesis class Q protein) (PIG-Q) (N-acetylglucosamyl transferase component GPI1). | |||||
|
PLXA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.027068 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70206, Q5DTR0 | Gene names | Plxna1, KIAA4053 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Plexin-1) (Plex 1). | |||||
|
TIEG3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.000368 (rank : 28) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K1S5, Q8BI37 | Gene names | Tieg3, Tieg2b | |||
|
Domain Architecture |

|
|||||
| Description | Transforming growth factor-beta-inducible early growth response protein 3 (TGFB-inducible early growth response protein 3) (TIEG-3) (TGFB-inducible early growth response protein 2b). | |||||
|
BCAR3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.018813 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75815, Q6UW40, Q9BR50 | Gene names | BCAR3, NSP2, SH2D3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast cancer anti-estrogen resistance protein 3 (SH2 domain- containing protein 3B) (Novel SH2-containing protein 2). | |||||
|
BCAR3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.019977 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QZK2, Q3TNC9, Q3UP10 | Gene names | Bcar3, And34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast cancer anti-estrogen resistance protein 3 (p130Cas-binding protein AND-34). | |||||
|
DHE4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.008583 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49448, Q9UDQ4 | Gene names | GLUD2, GLUDP1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate dehydrogenase 2, mitochondrial precursor (EC 1.4.1.3) (GDH). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.003141 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
SATB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.007187 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60611 | Gene names | Satb1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB1 (Special AT-rich sequence-binding protein 1). | |||||
|
ZIMP7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.014351 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NF64, O94790, Q659A8, Q6JKL5, Q8WTX8, Q96Q01, Q9BQH7 | Gene names | ZIMP7, KIAA1886 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
CG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.011220 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
IRK10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.015091 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JM63 | Gene names | Kcnj10 | |||
|
Domain Architecture |
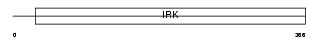
|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 10 (Potassium channel, inwardly rectifying subfamily J member 10) (Inward rectifier K(+) channel Kir4.1). | |||||
|
T22D4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.008432 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQN3 | Gene names | Tsc22d4, Tilz2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2). | |||||
|
YTHD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.060946 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BYK6, Q3UVI5, Q6NXJ6, Q6NXJ8, Q8BKB6 | Gene names | Ythdf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain family protein 3. | |||||
|
COE4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BQW3, Q5JY53, Q9NUB6, Q9P2A6 | Gene names | EBF4, COE4, KIAA1442 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE4 (Early B-cell factor 4) (EBF-4) (Olf-1/EBF- like 4) (OE-4) (O/E-4). | |||||
|
COE4_MOUSE
|
||||||
| NC score | 0.997630 (rank : 2) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K4J2, Q8K4J1, Q8K4J3, Q8K4J4, Q8K4J5 | Gene names | Ebf4, Coe4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE4 (Early B-cell factor 4) (EBF-4) (Olf-1/EBF- like 4) (OE-4) (O/E-4). | |||||
|
COE2_MOUSE
|
||||||
| NC score | 0.994740 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08792, Q543D5 | Gene names | Ebf2, Coe2, Mmot1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE2 (Early B-cell factor 2) (EBF-2) (Olf-1/EBF- like 3) (OE-3) (O/E-3) (Metencephalon-mesencephalon-olfactory transcription factor 1) (MET-mesencephalon-olfactory transcription factor 1) (MET-mesencephalon-olfactory TF1). | |||||
|
COE1_HUMAN
|
||||||
| NC score | 0.993607 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UH73, Q8IW11 | Gene names | EBF1, COE1, EBF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
COE1_MOUSE
|
||||||
| NC score | 0.993607 (rank : 5) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07802 | Gene names | Ebf1, Coe1, Ebf | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
COE3_MOUSE
|
||||||
| NC score | 0.991308 (rank : 6) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08791, O08793 | Gene names | Ebf3, Coe3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
COE3_HUMAN
|
||||||
| NC score | 0.991072 (rank : 7) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H4W6, Q5T6H9, Q9H4W5 | Gene names | EBF3, COE3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
YTHD3_MOUSE
|
||||||
| NC score | 0.060946 (rank : 8) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BYK6, Q3UVI5, Q6NXJ6, Q6NXJ8, Q8BKB6 | Gene names | Ythdf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain family protein 3. | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.046086 (rank : 9) | θ value | 0.0113563 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
PLXA1_MOUSE
|
||||||
| NC score | 0.027068 (rank : 10) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70206, Q5DTR0 | Gene names | Plxna1, KIAA4053 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Plexin-1) (Plex 1). | |||||
|
PIGQ_HUMAN
|
||||||
| NC score | 0.020555 (rank : 11) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BRB3, O14927, Q96G00, Q96S22, Q9UJH4 | Gene names | PIGQ, GPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q (EC 2.4.1.198) (Phosphatidylinositol-glycan biosynthesis class Q protein) (PIG-Q) (N-acetylglucosamyl transferase component GPI1). | |||||
|
BCAR3_MOUSE
|
||||||
| NC score | 0.019977 (rank : 12) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QZK2, Q3TNC9, Q3UP10 | Gene names | Bcar3, And34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast cancer anti-estrogen resistance protein 3 (p130Cas-binding protein AND-34). | |||||
|
IRK10_HUMAN
|
||||||
| NC score | 0.019543 (rank : 13) | θ value | 5.27518 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78508, Q8N4I7, Q92808 | Gene names | KCNJ10 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 10 (Potassium channel, inwardly rectifying subfamily J member 10) (Inward rectifier K(+) channel Kir1.2) (ATP-dependent inwardly rectifying potassium channel Kir4.1). | |||||
|
CCNL2_HUMAN
|
||||||
| NC score | 0.019143 (rank : 14) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96S94, Q5T2N5, Q5T2N6, Q6IQ12, Q7Z4Z8, Q8N3C9, Q8N3D5, Q8NHE3, Q8TEL0, Q96B00 | Gene names | CCNL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Paneth cell-enhanced expression protein). | |||||
|
BCAR3_HUMAN
|
||||||
| NC score | 0.018813 (rank : 15) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75815, Q6UW40, Q9BR50 | Gene names | BCAR3, NSP2, SH2D3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast cancer anti-estrogen resistance protein 3 (SH2 domain- containing protein 3B) (Novel SH2-containing protein 2). | |||||
|
UBQL3_HUMAN
|
||||||
| NC score | 0.018338 (rank : 16) | θ value | 0.62314 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H347, Q9NRE0 | Gene names | UBQLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.015390 (rank : 17) | θ value | 1.38821 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
IRK10_MOUSE
|
||||||
| NC score | 0.015091 (rank : 18) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JM63 | Gene names | Kcnj10 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-sensitive inward rectifier potassium channel 10 (Potassium channel, inwardly rectifying subfamily J member 10) (Inward rectifier K(+) channel Kir4.1). | |||||
|
ZIMP7_HUMAN
|
||||||
| NC score | 0.014351 (rank : 19) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NF64, O94790, Q659A8, Q6JKL5, Q8WTX8, Q96Q01, Q9BQH7 | Gene names | ZIMP7, KIAA1886 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.013347 (rank : 20) | θ value | 0.279714 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
CG1_HUMAN
|
||||||
| NC score | 0.011220 (rank : 21) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
ASTL_HUMAN
|
||||||
| NC score | 0.010262 (rank : 22) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
DHE4_HUMAN
|
||||||
| NC score | 0.008583 (rank : 23) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49448, Q9UDQ4 | Gene names | GLUD2, GLUDP1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate dehydrogenase 2, mitochondrial precursor (EC 1.4.1.3) (GDH). | |||||
|
T22D4_MOUSE
|
||||||
| NC score | 0.008432 (rank : 24) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQN3 | Gene names | Tsc22d4, Tilz2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2). | |||||
|
SATB1_MOUSE
|
||||||
| NC score | 0.007187 (rank : 25) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60611 | Gene names | Satb1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB1 (Special AT-rich sequence-binding protein 1). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.003141 (rank : 26) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
GLI1_HUMAN
|
||||||
| NC score | 0.001686 (rank : 27) | θ value | 1.81305 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||
|
TIEG3_MOUSE
|
||||||
| NC score | 0.000368 (rank : 28) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K1S5, Q8BI37 | Gene names | Tieg3, Tieg2b | |||
|
Domain Architecture |

|
|||||
| Description | Transforming growth factor-beta-inducible early growth response protein 3 (TGFB-inducible early growth response protein 3) (TIEG-3) (TGFB-inducible early growth response protein 2b). | |||||