Please be patient as the page loads
|
CHST8_MOUSE
|
||||||
| SwissProt Accessions | Q8BQ86, Q76EC6, Q80XD4 | Gene names | Chst8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 8 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 1) (GalNAc-4-O-sulfotransferase 1) (GalNAc-4-ST1) (GalNAc4ST-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CHST8_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998353 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H2A9, Q9H3N2 | Gene names | CHST8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 8 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 1) (GalNAc-4-O-sulfotransferase 1) (GalNAc-4-ST1) (GalNAc4ST-1). | |||||
|
CHST8_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BQ86, Q76EC6, Q80XD4 | Gene names | Chst8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 8 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 1) (GalNAc-4-O-sulfotransferase 1) (GalNAc-4-ST1) (GalNAc4ST-1). | |||||
|
CHST9_HUMAN
|
||||||
| θ value | 6.24247e-110 (rank : 3) | NC score | 0.984728 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7L1S5, Q6UX69, Q9BXH3, Q9BXH4, Q9BZW9 | Gene names | CHST9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 9 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 2) (GalNAc-4-O-sulfotransferase 2) (GalNAc-4-ST2). | |||||
|
CHST9_MOUSE
|
||||||
| θ value | 1.87955e-106 (rank : 4) | NC score | 0.985928 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q76EC5 | Gene names | Chst9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 9 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 2) (GalNAc-4-O-sulfotransferase 2) (GalNAc-4-ST2). | |||||
|
CHSTB_MOUSE
|
||||||
| θ value | 9.45883e-58 (rank : 5) | NC score | 0.944957 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JME2, Q9JJS2 | Gene names | Chst11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 11 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 1) (Chondroitin 4-sulfotransferase 1) (C4ST) (C4ST-1) (C4S-1). | |||||
|
CHSTB_HUMAN
|
||||||
| θ value | 6.13105e-57 (rank : 6) | NC score | 0.944786 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NPF2, Q9NXY6, Q9NY36 | Gene names | CHST11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 11 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 1) (Chondroitin 4-sulfotransferase 1) (C4ST) (C4ST-1) (C4S-1). | |||||
|
CHSTC_HUMAN
|
||||||
| θ value | 9.18288e-45 (rank : 7) | NC score | 0.922137 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NRB3, Q9NXY7 | Gene names | CHST12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 12 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 2) (Chondroitin 4-sulfotransferase 2) (C4ST2) (C4ST- 2) (Sulfotransferase Hlo). | |||||
|
CHSTD_HUMAN
|
||||||
| θ value | 1.32601e-43 (rank : 8) | NC score | 0.945256 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NET6 | Gene names | CHST13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 13 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 3) (Chondroitin 4-sulfotransferase 3) (C4ST3) (C4ST- 3). | |||||
|
CHSTC_MOUSE
|
||||||
| θ value | 3.85808e-43 (rank : 9) | NC score | 0.917873 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99LL3, Q9JJS3 | Gene names | Chst12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 12 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 2) (Chondroitin 4-sulfotransferase 2) (C4ST2) (C4ST- 2). | |||||
|
CHSTE_MOUSE
|
||||||
| θ value | 8.89434e-40 (rank : 10) | NC score | 0.910277 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80V53, Q3TXA1, Q8R304, Q9D0P2 | Gene names | D4st1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase D4ST1 (EC 2.8.2.-) (Dermatan 4- sulfotransferase 1) (D4ST-1). | |||||
|
CHSTE_HUMAN
|
||||||
| θ value | 9.83387e-39 (rank : 11) | NC score | 0.909963 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NCH0, Q6PJ31, Q6UXA0, Q96P94 | Gene names | D4ST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase D4ST1 (EC 2.8.2.-) (Dermatan 4- sulfotransferase 1) (D4ST-1) (hD4ST). | |||||
|
CHSTA_MOUSE
|
||||||
| θ value | 2.19075e-38 (rank : 12) | NC score | 0.906179 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6PGK7, Q8BKU3, Q8BL08, Q8R2Y5, Q91Y40 | Gene names | Chst10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 10 (EC 2.8.2.-) (HNK-1 sulfotransferase) (HNK1ST) (HNK-1ST). | |||||
|
CHSTA_HUMAN
|
||||||
| θ value | 2.42216e-37 (rank : 13) | NC score | 0.904470 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43529 | Gene names | CHST10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 10 (EC 2.8.2.-) (HNK-1 sulfotransferase) (HNK1ST) (HNK-1ST) (huHNK-1ST). | |||||
|
RIP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.034221 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CWY9 | Gene names | Rpain, Rip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RPA-interacting protein. | |||||
|
ALK_MOUSE
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.000294 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97793 | Gene names | Alk | |||
|
Domain Architecture |

|
|||||
| Description | ALK tyrosine kinase receptor precursor (EC 2.7.10.1) (Anaplastic lymphoma kinase) (CD246 antigen). | |||||
|
GRIK5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.008373 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
RUSC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.013645 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
CN120_MOUSE
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.017049 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DB96, Q8CIJ2 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C14orf120 homolog. | |||||
|
GRIK5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.007678 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
PJA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.009867 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55176, Q8CFU2, Q99MJ1, Q99MJ2, Q99MJ3, Q9DB04 | Gene names | Pja1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin protein ligase Praja1 (EC 6.3.2.-). | |||||
|
TES_MOUSE
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.013256 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47226 | Gene names | Tes | |||
|
Domain Architecture |
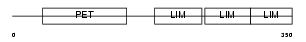
|
|||||
| Description | Testin (TES1/TES2). | |||||
|
CREB5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.008484 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.004998 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
IF4G1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.006308 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | -0.000494 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
ADA2B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | -0.001349 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 891 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30545 | Gene names | Adra2b | |||
|
Domain Architecture |
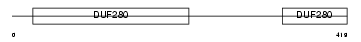
|
|||||
| Description | Alpha-2B adrenergic receptor (Alpha-2B adrenoceptor) (Alpha-2B adrenoreceptor). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.004668 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.003095 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.002423 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CHST8_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BQ86, Q76EC6, Q80XD4 | Gene names | Chst8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 8 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 1) (GalNAc-4-O-sulfotransferase 1) (GalNAc-4-ST1) (GalNAc4ST-1). | |||||
|
CHST8_HUMAN
|
||||||
| NC score | 0.998353 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H2A9, Q9H3N2 | Gene names | CHST8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 8 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 1) (GalNAc-4-O-sulfotransferase 1) (GalNAc-4-ST1) (GalNAc4ST-1). | |||||
|
CHST9_MOUSE
|
||||||
| NC score | 0.985928 (rank : 3) | θ value | 1.87955e-106 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q76EC5 | Gene names | Chst9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 9 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 2) (GalNAc-4-O-sulfotransferase 2) (GalNAc-4-ST2). | |||||
|
CHST9_HUMAN
|
||||||
| NC score | 0.984728 (rank : 4) | θ value | 6.24247e-110 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7L1S5, Q6UX69, Q9BXH3, Q9BXH4, Q9BZW9 | Gene names | CHST9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 9 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 2) (GalNAc-4-O-sulfotransferase 2) (GalNAc-4-ST2). | |||||
|
CHSTD_HUMAN
|
||||||
| NC score | 0.945256 (rank : 5) | θ value | 1.32601e-43 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NET6 | Gene names | CHST13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 13 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 3) (Chondroitin 4-sulfotransferase 3) (C4ST3) (C4ST- 3). | |||||
|
CHSTB_MOUSE
|
||||||
| NC score | 0.944957 (rank : 6) | θ value | 9.45883e-58 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JME2, Q9JJS2 | Gene names | Chst11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 11 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 1) (Chondroitin 4-sulfotransferase 1) (C4ST) (C4ST-1) (C4S-1). | |||||
|
CHSTB_HUMAN
|
||||||
| NC score | 0.944786 (rank : 7) | θ value | 6.13105e-57 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NPF2, Q9NXY6, Q9NY36 | Gene names | CHST11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 11 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 1) (Chondroitin 4-sulfotransferase 1) (C4ST) (C4ST-1) (C4S-1). | |||||
|
CHSTC_HUMAN
|
||||||
| NC score | 0.922137 (rank : 8) | θ value | 9.18288e-45 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NRB3, Q9NXY7 | Gene names | CHST12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 12 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 2) (Chondroitin 4-sulfotransferase 2) (C4ST2) (C4ST- 2) (Sulfotransferase Hlo). | |||||
|
CHSTC_MOUSE
|
||||||
| NC score | 0.917873 (rank : 9) | θ value | 3.85808e-43 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99LL3, Q9JJS3 | Gene names | Chst12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 12 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 2) (Chondroitin 4-sulfotransferase 2) (C4ST2) (C4ST- 2). | |||||
|
CHSTE_MOUSE
|
||||||
| NC score | 0.910277 (rank : 10) | θ value | 8.89434e-40 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80V53, Q3TXA1, Q8R304, Q9D0P2 | Gene names | D4st1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase D4ST1 (EC 2.8.2.-) (Dermatan 4- sulfotransferase 1) (D4ST-1). | |||||
|
CHSTE_HUMAN
|
||||||
| NC score | 0.909963 (rank : 11) | θ value | 9.83387e-39 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NCH0, Q6PJ31, Q6UXA0, Q96P94 | Gene names | D4ST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase D4ST1 (EC 2.8.2.-) (Dermatan 4- sulfotransferase 1) (D4ST-1) (hD4ST). | |||||
|
CHSTA_MOUSE
|
||||||
| NC score | 0.906179 (rank : 12) | θ value | 2.19075e-38 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6PGK7, Q8BKU3, Q8BL08, Q8R2Y5, Q91Y40 | Gene names | Chst10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 10 (EC 2.8.2.-) (HNK-1 sulfotransferase) (HNK1ST) (HNK-1ST). | |||||
|
CHSTA_HUMAN
|
||||||
| NC score | 0.904470 (rank : 13) | θ value | 2.42216e-37 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43529 | Gene names | CHST10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 10 (EC 2.8.2.-) (HNK-1 sulfotransferase) (HNK1ST) (HNK-1ST) (huHNK-1ST). | |||||
|
RIP_MOUSE
|
||||||
| NC score | 0.034221 (rank : 14) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CWY9 | Gene names | Rpain, Rip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RPA-interacting protein. | |||||
|
CN120_MOUSE
|
||||||
| NC score | 0.017049 (rank : 15) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DB96, Q8CIJ2 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C14orf120 homolog. | |||||
|
RUSC2_MOUSE
|
||||||
| NC score | 0.013645 (rank : 16) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
TES_MOUSE
|
||||||
| NC score | 0.013256 (rank : 17) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47226 | Gene names | Tes | |||
|
Domain Architecture |

|
|||||
| Description | Testin (TES1/TES2). | |||||
|
PJA1_MOUSE
|
||||||
| NC score | 0.009867 (rank : 18) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55176, Q8CFU2, Q99MJ1, Q99MJ2, Q99MJ3, Q9DB04 | Gene names | Pja1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin protein ligase Praja1 (EC 6.3.2.-). | |||||
|
CREB5_HUMAN
|
||||||
| NC score | 0.008484 (rank : 19) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
GRIK5_HUMAN
|
||||||
| NC score | 0.008373 (rank : 20) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
GRIK5_MOUSE
|
||||||
| NC score | 0.007678 (rank : 21) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
IF4G1_HUMAN
|
||||||
| NC score | 0.006308 (rank : 22) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.004998 (rank : 23) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.004668 (rank : 24) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.003095 (rank : 25) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.002423 (rank : 26) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ALK_MOUSE
|
||||||
| NC score | 0.000294 (rank : 27) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97793 | Gene names | Alk | |||
|
Domain Architecture |

|
|||||
| Description | ALK tyrosine kinase receptor precursor (EC 2.7.10.1) (Anaplastic lymphoma kinase) (CD246 antigen). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | -0.000494 (rank : 28) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
ADA2B_MOUSE
|
||||||
| NC score | -0.001349 (rank : 29) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 891 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30545 | Gene names | Adra2b | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2B adrenergic receptor (Alpha-2B adrenoceptor) (Alpha-2B adrenoreceptor). | |||||