Please be patient as the page loads
|
CHSTA_MOUSE
|
||||||
| SwissProt Accessions | Q6PGK7, Q8BKU3, Q8BL08, Q8R2Y5, Q91Y40 | Gene names | Chst10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 10 (EC 2.8.2.-) (HNK-1 sulfotransferase) (HNK1ST) (HNK-1ST). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CHSTA_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998863 (rank : 2) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43529 | Gene names | CHST10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 10 (EC 2.8.2.-) (HNK-1 sulfotransferase) (HNK1ST) (HNK-1ST) (huHNK-1ST). | |||||
|
CHSTA_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PGK7, Q8BKU3, Q8BL08, Q8R2Y5, Q91Y40 | Gene names | Chst10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 10 (EC 2.8.2.-) (HNK-1 sulfotransferase) (HNK1ST) (HNK-1ST). | |||||
|
CHST9_HUMAN
|
||||||
| θ value | 3.99248e-40 (rank : 3) | NC score | 0.905898 (rank : 8) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7L1S5, Q6UX69, Q9BXH3, Q9BXH4, Q9BZW9 | Gene names | CHST9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 9 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 2) (GalNAc-4-O-sulfotransferase 2) (GalNAc-4-ST2). | |||||
|
CHSTB_MOUSE
|
||||||
| θ value | 1.6774e-38 (rank : 4) | NC score | 0.909711 (rank : 4) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JME2, Q9JJS2 | Gene names | Chst11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 11 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 1) (Chondroitin 4-sulfotransferase 1) (C4ST) (C4ST-1) (C4S-1). | |||||
|
CHST8_MOUSE
|
||||||
| θ value | 2.19075e-38 (rank : 5) | NC score | 0.906179 (rank : 7) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BQ86, Q76EC6, Q80XD4 | Gene names | Chst8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 8 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 1) (GalNAc-4-O-sulfotransferase 1) (GalNAc-4-ST1) (GalNAc4ST-1). | |||||
|
CHSTB_HUMAN
|
||||||
| θ value | 8.32485e-38 (rank : 6) | NC score | 0.908991 (rank : 5) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NPF2, Q9NXY6, Q9NY36 | Gene names | CHST11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 11 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 1) (Chondroitin 4-sulfotransferase 1) (C4ST) (C4ST-1) (C4S-1). | |||||
|
CHST8_HUMAN
|
||||||
| θ value | 2.42216e-37 (rank : 7) | NC score | 0.906203 (rank : 6) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H2A9, Q9H3N2 | Gene names | CHST8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 8 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 1) (GalNAc-4-O-sulfotransferase 1) (GalNAc-4-ST1) (GalNAc4ST-1). | |||||
|
CHST9_MOUSE
|
||||||
| θ value | 4.13158e-37 (rank : 8) | NC score | 0.904747 (rank : 9) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q76EC5 | Gene names | Chst9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 9 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 2) (GalNAc-4-O-sulfotransferase 2) (GalNAc-4-ST2). | |||||
|
CHSTC_MOUSE
|
||||||
| θ value | 1.79631e-32 (rank : 9) | NC score | 0.888530 (rank : 10) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99LL3, Q9JJS3 | Gene names | Chst12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 12 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 2) (Chondroitin 4-sulfotransferase 2) (C4ST2) (C4ST- 2). | |||||
|
CHSTD_HUMAN
|
||||||
| θ value | 3.3877e-31 (rank : 10) | NC score | 0.910967 (rank : 3) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NET6 | Gene names | CHST13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 13 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 3) (Chondroitin 4-sulfotransferase 3) (C4ST3) (C4ST- 3). | |||||
|
CHSTC_HUMAN
|
||||||
| θ value | 4.42448e-31 (rank : 11) | NC score | 0.887733 (rank : 11) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NRB3, Q9NXY7 | Gene names | CHST12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 12 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 2) (Chondroitin 4-sulfotransferase 2) (C4ST2) (C4ST- 2) (Sulfotransferase Hlo). | |||||
|
CHSTE_MOUSE
|
||||||
| θ value | 5.59698e-26 (rank : 12) | NC score | 0.870787 (rank : 12) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80V53, Q3TXA1, Q8R304, Q9D0P2 | Gene names | D4st1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase D4ST1 (EC 2.8.2.-) (Dermatan 4- sulfotransferase 1) (D4ST-1). | |||||
|
CHSTE_HUMAN
|
||||||
| θ value | 3.62785e-25 (rank : 13) | NC score | 0.869019 (rank : 13) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NCH0, Q6PJ31, Q6UXA0, Q96P94 | Gene names | D4ST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase D4ST1 (EC 2.8.2.-) (Dermatan 4- sulfotransferase 1) (D4ST-1) (hD4ST). | |||||
|
EXOC6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.021996 (rank : 16) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R313 | Gene names | Exoc6, Sec15a, Sec15l1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 6 (Exocyst complex component Sec15A) (Sec15- like 1). | |||||
|
KPRB_MOUSE
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.028865 (rank : 14) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R574 | Gene names | Prpsap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoribosyl pyrophosphate synthetase-associated protein 2 (PRPP synthetase-associated protein 2) (41 kDa phosphoribosypyrophosphate synthetase-associated protein) (PAP41). | |||||
|
KLH11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.013141 (rank : 21) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NVR0 | Gene names | KLHL11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 11 precursor. | |||||
|
KLH11_MOUSE
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.013144 (rank : 20) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CE33, Q3TDZ6, Q91VP6 | Gene names | Klhl11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 11 precursor. | |||||
|
ASPM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.012062 (rank : 22) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CJ27, O88482, Q8BJI8, Q8BKT4 | Gene names | Aspm, Calmbp1, Sha1 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein homolog (Calmodulin-binding protein 1) (Spindle and hydroxyurea checkpoint abnormal protein) (Calmodulin-binding protein Sha1). | |||||
|
LBC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.016455 (rank : 19) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12802 | Gene names | LBC | |||
|
Domain Architecture |
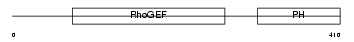
|
|||||
| Description | LBC oncogene (P47) (Lymphoid blast crisis oncogene). | |||||
|
KPRA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.022093 (rank : 15) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14558, Q96H06 | Gene names | PRPSAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoribosyl pyrophosphate synthetase-associated protein 1 (PRPP synthetase-associated protein 1) (39 kDa phosphoribosypyrophosphate synthetase-associated protein) (PAP39). | |||||
|
KPRA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.020590 (rank : 17) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D0M1 | Gene names | Prpsap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoribosyl pyrophosphate synthetase-associated protein 1 (PRPP synthetase-associated protein 1) (39 kDa phosphoribosypyrophosphate synthetase-associated protein) (PAP39). | |||||
|
KPRB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.020549 (rank : 18) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60256 | Gene names | PRPSAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoribosyl pyrophosphate synthetase-associated protein 2 (PRPP synthetase-associated protein 2) (41 kDa phosphoribosypyrophosphate synthetase-associated protein) (PAP41). | |||||
|
MET_MOUSE
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | -0.001489 (rank : 23) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P16056, Q62125 | Gene names | Met | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor receptor precursor (EC 2.7.10.1) (HGF receptor) (Scatter factor receptor) (SF receptor) (HGF/SF receptor) (Met proto-oncogene tyrosine kinase) (c-Met). | |||||
|
CHSTA_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PGK7, Q8BKU3, Q8BL08, Q8R2Y5, Q91Y40 | Gene names | Chst10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 10 (EC 2.8.2.-) (HNK-1 sulfotransferase) (HNK1ST) (HNK-1ST). | |||||
|
CHSTA_HUMAN
|
||||||
| NC score | 0.998863 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43529 | Gene names | CHST10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 10 (EC 2.8.2.-) (HNK-1 sulfotransferase) (HNK1ST) (HNK-1ST) (huHNK-1ST). | |||||
|
CHSTD_HUMAN
|
||||||
| NC score | 0.910967 (rank : 3) | θ value | 3.3877e-31 (rank : 10) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NET6 | Gene names | CHST13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 13 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 3) (Chondroitin 4-sulfotransferase 3) (C4ST3) (C4ST- 3). | |||||
|
CHSTB_MOUSE
|
||||||
| NC score | 0.909711 (rank : 4) | θ value | 1.6774e-38 (rank : 4) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JME2, Q9JJS2 | Gene names | Chst11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 11 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 1) (Chondroitin 4-sulfotransferase 1) (C4ST) (C4ST-1) (C4S-1). | |||||
|
CHSTB_HUMAN
|
||||||
| NC score | 0.908991 (rank : 5) | θ value | 8.32485e-38 (rank : 6) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NPF2, Q9NXY6, Q9NY36 | Gene names | CHST11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 11 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 1) (Chondroitin 4-sulfotransferase 1) (C4ST) (C4ST-1) (C4S-1). | |||||
|
CHST8_HUMAN
|
||||||
| NC score | 0.906203 (rank : 6) | θ value | 2.42216e-37 (rank : 7) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H2A9, Q9H3N2 | Gene names | CHST8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 8 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 1) (GalNAc-4-O-sulfotransferase 1) (GalNAc-4-ST1) (GalNAc4ST-1). | |||||
|
CHST8_MOUSE
|
||||||
| NC score | 0.906179 (rank : 7) | θ value | 2.19075e-38 (rank : 5) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BQ86, Q76EC6, Q80XD4 | Gene names | Chst8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 8 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 1) (GalNAc-4-O-sulfotransferase 1) (GalNAc-4-ST1) (GalNAc4ST-1). | |||||
|
CHST9_HUMAN
|
||||||
| NC score | 0.905898 (rank : 8) | θ value | 3.99248e-40 (rank : 3) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7L1S5, Q6UX69, Q9BXH3, Q9BXH4, Q9BZW9 | Gene names | CHST9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 9 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 2) (GalNAc-4-O-sulfotransferase 2) (GalNAc-4-ST2). | |||||
|
CHST9_MOUSE
|
||||||
| NC score | 0.904747 (rank : 9) | θ value | 4.13158e-37 (rank : 8) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q76EC5 | Gene names | Chst9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 9 (EC 2.8.2.-) (N-acetylgalactosamine-4- O-sulfotransferase 2) (GalNAc-4-O-sulfotransferase 2) (GalNAc-4-ST2). | |||||
|
CHSTC_MOUSE
|
||||||
| NC score | 0.888530 (rank : 10) | θ value | 1.79631e-32 (rank : 9) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99LL3, Q9JJS3 | Gene names | Chst12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 12 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 2) (Chondroitin 4-sulfotransferase 2) (C4ST2) (C4ST- 2). | |||||
|
CHSTC_HUMAN
|
||||||
| NC score | 0.887733 (rank : 11) | θ value | 4.42448e-31 (rank : 11) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NRB3, Q9NXY7 | Gene names | CHST12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase 12 (EC 2.8.2.5) (Chondroitin 4-O- sulfotransferase 2) (Chondroitin 4-sulfotransferase 2) (C4ST2) (C4ST- 2) (Sulfotransferase Hlo). | |||||
|
CHSTE_MOUSE
|
||||||
| NC score | 0.870787 (rank : 12) | θ value | 5.59698e-26 (rank : 12) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80V53, Q3TXA1, Q8R304, Q9D0P2 | Gene names | D4st1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase D4ST1 (EC 2.8.2.-) (Dermatan 4- sulfotransferase 1) (D4ST-1). | |||||
|
CHSTE_HUMAN
|
||||||
| NC score | 0.869019 (rank : 13) | θ value | 3.62785e-25 (rank : 13) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NCH0, Q6PJ31, Q6UXA0, Q96P94 | Gene names | D4ST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbohydrate sulfotransferase D4ST1 (EC 2.8.2.-) (Dermatan 4- sulfotransferase 1) (D4ST-1) (hD4ST). | |||||
|
KPRB_MOUSE
|
||||||
| NC score | 0.028865 (rank : 14) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R574 | Gene names | Prpsap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoribosyl pyrophosphate synthetase-associated protein 2 (PRPP synthetase-associated protein 2) (41 kDa phosphoribosypyrophosphate synthetase-associated protein) (PAP41). | |||||
|
KPRA_HUMAN
|
||||||
| NC score | 0.022093 (rank : 15) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14558, Q96H06 | Gene names | PRPSAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoribosyl pyrophosphate synthetase-associated protein 1 (PRPP synthetase-associated protein 1) (39 kDa phosphoribosypyrophosphate synthetase-associated protein) (PAP39). | |||||
|
EXOC6_MOUSE
|
||||||
| NC score | 0.021996 (rank : 16) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R313 | Gene names | Exoc6, Sec15a, Sec15l1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 6 (Exocyst complex component Sec15A) (Sec15- like 1). | |||||
|
KPRA_MOUSE
|
||||||
| NC score | 0.020590 (rank : 17) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D0M1 | Gene names | Prpsap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoribosyl pyrophosphate synthetase-associated protein 1 (PRPP synthetase-associated protein 1) (39 kDa phosphoribosypyrophosphate synthetase-associated protein) (PAP39). | |||||
|
KPRB_HUMAN
|
||||||
| NC score | 0.020549 (rank : 18) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60256 | Gene names | PRPSAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoribosyl pyrophosphate synthetase-associated protein 2 (PRPP synthetase-associated protein 2) (41 kDa phosphoribosypyrophosphate synthetase-associated protein) (PAP41). | |||||
|
LBC_HUMAN
|
||||||
| NC score | 0.016455 (rank : 19) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12802 | Gene names | LBC | |||
|
Domain Architecture |

|
|||||
| Description | LBC oncogene (P47) (Lymphoid blast crisis oncogene). | |||||
|
KLH11_MOUSE
|
||||||
| NC score | 0.013144 (rank : 20) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CE33, Q3TDZ6, Q91VP6 | Gene names | Klhl11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 11 precursor. | |||||
|
KLH11_HUMAN
|
||||||
| NC score | 0.013141 (rank : 21) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NVR0 | Gene names | KLHL11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 11 precursor. | |||||
|
ASPM_MOUSE
|
||||||
| NC score | 0.012062 (rank : 22) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CJ27, O88482, Q8BJI8, Q8BKT4 | Gene names | Aspm, Calmbp1, Sha1 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein homolog (Calmodulin-binding protein 1) (Spindle and hydroxyurea checkpoint abnormal protein) (Calmodulin-binding protein Sha1). | |||||
|
MET_MOUSE
|
||||||
| NC score | -0.001489 (rank : 23) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P16056, Q62125 | Gene names | Met | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor receptor precursor (EC 2.7.10.1) (HGF receptor) (Scatter factor receptor) (SF receptor) (HGF/SF receptor) (Met proto-oncogene tyrosine kinase) (c-Met). | |||||