Please be patient as the page loads
|
CDCP1_MOUSE
|
||||||
| SwissProt Accessions | Q5U462, Q810J0, Q921M9 | Gene names | Cdcp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB domain-containing protein 1 precursor (Transmembrane and associated with src kinases) (Membrane glycoprotein gp140) (CD318 antigen). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CDCP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.990878 (rank : 2) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H5V8, Q49UB4, Q6NT71, Q6U9Y2, Q8WU91, Q96QU7, Q9H676, Q9H8C2 | Gene names | CDCP1, TRASK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB domain-containing protein 1 precursor (Transmembrane and associated with src kinases) (Membrane glycoprotein gp140) (Subtractive immunization M plus HEp3-associated 135 kDa protein) (SIMA135) (CD318 antigen). | |||||
|
CDCP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5U462, Q810J0, Q921M9 | Gene names | Cdcp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB domain-containing protein 1 precursor (Transmembrane and associated with src kinases) (Membrane glycoprotein gp140) (CD318 antigen). | |||||
|
MASP1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 3) | NC score | 0.052193 (rank : 18) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P98064 | Gene names | Masp1, Crarf | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) [Contains: Complement- activating component of Ra-reactive factor heavy chain; Complement- activating component of Ra-reactive factor light chain]. | |||||
|
MASP1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 4) | NC score | 0.052360 (rank : 17) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||
|
CADH2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 5) | NC score | 0.022454 (rank : 25) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15116, Q64260 | Gene names | Cdh2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-2 precursor (Neural-cadherin) (N-cadherin) (CDw325 antigen). | |||||
|
CSMD2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 6) | NC score | 0.071403 (rank : 3) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.47712 (rank : 7) | NC score | 0.016804 (rank : 27) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CSMD1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 8) | NC score | 0.067609 (rank : 6) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q923L3, Q8BUV1, Q8BYQ3 | Gene names | Csmd1 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
MFRP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 9) | NC score | 0.070851 (rank : 4) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
EXOC7_HUMAN
|
||||||
| θ value | 1.06291 (rank : 10) | NC score | 0.057375 (rank : 14) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPT5, Q8ND93, Q8WV91, Q96FF0, Q9H8C3, Q9H9X3, Q9HA32 | Gene names | EXOC7, EXO70, KIAA1067 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 7 (Exocyst complex component Exo70). | |||||
|
EXOC7_MOUSE
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.057508 (rank : 11) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35250, Q8K121 | Gene names | Exoc7, Exo70 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 7 (Exocyst complex component Exo70). | |||||
|
CSMD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.062287 (rank : 8) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
CUBN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.040886 (rank : 20) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
DCBD2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.061321 (rank : 9) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
KDEL1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.027961 (rank : 23) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHP7, Q8BPU6, Q8C528, Q8R591, Q9D8P1 | Gene names | Kdelc1, Ep58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KDEL motif-containing protein 1 precursor (ER protein 58). | |||||
|
PDXL2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 16) | NC score | 0.034932 (rank : 21) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZ53, Q6UVY4, Q8WUV6 | Gene names | PODXL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
DCBD2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.057423 (rank : 13) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91ZV3, Q8BKI4 | Gene names | Dcbld2, Esdn | |||
|
Domain Architecture |
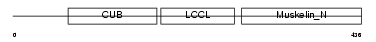
|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein). | |||||
|
MOV10_MOUSE
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.029605 (rank : 22) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23249, Q9DC64 | Gene names | Mov10, Gb110 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase MOV-10 (EC 3.6.1.-) (Moloney leukemia virus 10 protein). | |||||
|
SORC1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.016608 (rank : 28) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLC4, Q8VI45, Q922I1, Q9QY21 | Gene names | Sorcs1, Sorcs | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS1 precursor (mSorCS). | |||||
|
PCOC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.068910 (rank : 5) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
CSMD3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.059177 (rank : 10) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z407, Q96PZ3 | Gene names | CSMD3, KIAA1894 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 3 precursor (CUB and sushi multiple domains protein 3). | |||||
|
MOV10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.027328 (rank : 24) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCE1, Q8TEF0, Q9BSY3, Q9BUJ9 | Gene names | MOV10, KIAA1631 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase MOV-10 (EC 3.6.1.-) (Moloney leukemia virus 10 protein). | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.006852 (rank : 29) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
HNRPU_HUMAN
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.017827 (rank : 26) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00839, O75507, Q96HY9 | Gene names | HNRPU, SAFA, U21.1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U (hnRNP U) (Scaffold attachment factor A) (SAF-A) (p120) (pp120). | |||||
|
TLL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.057442 (rank : 12) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6L7, Q6PJN5, Q9UQ00 | Gene names | TLL2, KIAA0932 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TLL2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.055531 (rank : 15) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVM6 | Gene names | Tll2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
DMBT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.042478 (rank : 19) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
MAGI2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.005825 (rank : 32) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
SPB5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.005999 (rank : 30) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36952 | Gene names | SERPINB5, PI5 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B5 precursor (Maspin) (Protease inhibitor 5). | |||||
|
SPB5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.005987 (rank : 31) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70124 | Gene names | Serpinb5, Pi5, Spi5, Spi7 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B5 precursor (Maspin) (Protease inhibitor 5). | |||||
|
TLL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.054767 (rank : 16) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43897, Q96AN3, Q9NQS4 | Gene names | TLL1, TLL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-). | |||||
|
ZN710_MOUSE
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | -0.000205 (rank : 33) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3U288, Q3U341, Q6P9R3, Q8BJH0, Q8BTL6 | Gene names | Znf710, Zfp710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
PCOC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.064105 (rank : 7) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
CDCP1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5U462, Q810J0, Q921M9 | Gene names | Cdcp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB domain-containing protein 1 precursor (Transmembrane and associated with src kinases) (Membrane glycoprotein gp140) (CD318 antigen). | |||||
|
CDCP1_HUMAN
|
||||||
| NC score | 0.990878 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H5V8, Q49UB4, Q6NT71, Q6U9Y2, Q8WU91, Q96QU7, Q9H676, Q9H8C2 | Gene names | CDCP1, TRASK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB domain-containing protein 1 precursor (Transmembrane and associated with src kinases) (Membrane glycoprotein gp140) (Subtractive immunization M plus HEp3-associated 135 kDa protein) (SIMA135) (CD318 antigen). | |||||
|
CSMD2_HUMAN
|
||||||
| NC score | 0.071403 (rank : 3) | θ value | 0.21417 (rank : 6) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
MFRP_MOUSE
|
||||||
| NC score | 0.070851 (rank : 4) | θ value | 0.813845 (rank : 9) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K480, Q8BPP4 | Gene names | Mfrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane frizzled-related protein (Membrane-type frizzled-related protein). | |||||
|
PCOC1_MOUSE
|
||||||
| NC score | 0.068910 (rank : 5) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
CSMD1_MOUSE
|
||||||
| NC score | 0.067609 (rank : 6) | θ value | 0.47712 (rank : 8) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q923L3, Q8BUV1, Q8BYQ3 | Gene names | Csmd1 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
PCOC1_HUMAN
|
||||||
| NC score | 0.064105 (rank : 7) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15113, O14550 | Gene names | PCOLCE, PCPE1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein). | |||||
|
CSMD1_HUMAN
|
||||||
| NC score | 0.062287 (rank : 8) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96PZ7, Q96QU9, Q96RM4 | Gene names | CSMD1, KIAA1890 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 1 precursor (CUB and sushi multiple domains protein 1). | |||||
|
DCBD2_HUMAN
|
||||||
| NC score | 0.061321 (rank : 9) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
CSMD3_HUMAN
|
||||||
| NC score | 0.059177 (rank : 10) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z407, Q96PZ3 | Gene names | CSMD3, KIAA1894 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 3 precursor (CUB and sushi multiple domains protein 3). | |||||
|
EXOC7_MOUSE
|
||||||
| NC score | 0.057508 (rank : 11) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35250, Q8K121 | Gene names | Exoc7, Exo70 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 7 (Exocyst complex component Exo70). | |||||
|
TLL2_HUMAN
|
||||||
| NC score | 0.057442 (rank : 12) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6L7, Q6PJN5, Q9UQ00 | Gene names | TLL2, KIAA0932 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
DCBD2_MOUSE
|
||||||
| NC score | 0.057423 (rank : 13) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91ZV3, Q8BKI4 | Gene names | Dcbld2, Esdn | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein). | |||||
|
EXOC7_HUMAN
|
||||||
| NC score | 0.057375 (rank : 14) | θ value | 1.06291 (rank : 10) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPT5, Q8ND93, Q8WV91, Q96FF0, Q9H8C3, Q9H9X3, Q9HA32 | Gene names | EXOC7, EXO70, KIAA1067 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 7 (Exocyst complex component Exo70). | |||||
|
TLL2_MOUSE
|
||||||
| NC score | 0.055531 (rank : 15) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVM6 | Gene names | Tll2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 2 precursor (EC 3.4.24.-). | |||||
|
TLL1_HUMAN
|
||||||
| NC score | 0.054767 (rank : 16) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43897, Q96AN3, Q9NQS4 | Gene names | TLL1, TLL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tolloid-like protein 1 precursor (EC 3.4.24.-). | |||||
|
MASP1_HUMAN
|
||||||
| NC score | 0.052360 (rank : 17) | θ value | 0.0431538 (rank : 4) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||
|
MASP1_MOUSE
|
||||||
| NC score | 0.052193 (rank : 18) | θ value | 0.0330416 (rank : 3) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P98064 | Gene names | Masp1, Crarf | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) [Contains: Complement- activating component of Ra-reactive factor heavy chain; Complement- activating component of Ra-reactive factor light chain]. | |||||
|
DMBT1_MOUSE
|
||||||
| NC score | 0.042478 (rank : 19) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
CUBN_MOUSE
|
||||||
| NC score | 0.040886 (rank : 20) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
PDXL2_HUMAN
|
||||||
| NC score | 0.034932 (rank : 21) | θ value | 1.81305 (rank : 16) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZ53, Q6UVY4, Q8WUV6 | Gene names | PODXL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
MOV10_MOUSE
|
||||||
| NC score | 0.029605 (rank : 22) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23249, Q9DC64 | Gene names | Mov10, Gb110 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase MOV-10 (EC 3.6.1.-) (Moloney leukemia virus 10 protein). | |||||
|
KDEL1_MOUSE
|
||||||
| NC score | 0.027961 (rank : 23) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHP7, Q8BPU6, Q8C528, Q8R591, Q9D8P1 | Gene names | Kdelc1, Ep58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KDEL motif-containing protein 1 precursor (ER protein 58). | |||||
|
MOV10_HUMAN
|
||||||
| NC score | 0.027328 (rank : 24) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCE1, Q8TEF0, Q9BSY3, Q9BUJ9 | Gene names | MOV10, KIAA1631 | |||
|
Domain Architecture |

|
|||||
| Description | Putative helicase MOV-10 (EC 3.6.1.-) (Moloney leukemia virus 10 protein). | |||||
|
CADH2_MOUSE
|
||||||
| NC score | 0.022454 (rank : 25) | θ value | 0.0563607 (rank : 5) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15116, Q64260 | Gene names | Cdh2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-2 precursor (Neural-cadherin) (N-cadherin) (CDw325 antigen). | |||||
|
HNRPU_HUMAN
|
||||||
| NC score | 0.017827 (rank : 26) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q00839, O75507, Q96HY9 | Gene names | HNRPU, SAFA, U21.1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U (hnRNP U) (Scaffold attachment factor A) (SAF-A) (p120) (pp120). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.016804 (rank : 27) | θ value | 0.47712 (rank : 7) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
SORC1_MOUSE
|
||||||
| NC score | 0.016608 (rank : 28) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLC4, Q8VI45, Q922I1, Q9QY21 | Gene names | Sorcs1, Sorcs | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS1 precursor (mSorCS). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.006852 (rank : 29) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
SPB5_HUMAN
|
||||||
| NC score | 0.005999 (rank : 30) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36952 | Gene names | SERPINB5, PI5 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B5 precursor (Maspin) (Protease inhibitor 5). | |||||
|
SPB5_MOUSE
|
||||||
| NC score | 0.005987 (rank : 31) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70124 | Gene names | Serpinb5, Pi5, Spi5, Spi7 | |||
|
Domain Architecture |

|
|||||
| Description | Serpin B5 precursor (Maspin) (Protease inhibitor 5). | |||||
|
MAGI2_MOUSE
|
||||||
| NC score | 0.005825 (rank : 32) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
ZN710_MOUSE
|
||||||
| NC score | -0.000205 (rank : 33) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3U288, Q3U341, Q6P9R3, Q8BJH0, Q8BTL6 | Gene names | Znf710, Zfp710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||