Please be patient as the page loads
|
CD5R2_MOUSE
|
||||||
| SwissProt Accessions | O35926, O35277 | Gene names | Cdk5r2, Nck5ai | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 2 precursor (CDK5 activator 2) (Cyclin-dependent kinase 5 regulatory subunit 2) (P39) (P39I). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CD5R2_MOUSE
|
||||||
| θ value | 1.52337e-140 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O35926, O35277 | Gene names | Cdk5r2, Nck5ai | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 2 precursor (CDK5 activator 2) (Cyclin-dependent kinase 5 regulatory subunit 2) (P39) (P39I). | |||||
|
CD5R2_HUMAN
|
||||||
| θ value | 2.51683e-135 (rank : 2) | NC score | 0.985870 (rank : 2) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13319, Q4ZFW6 | Gene names | CDK5R2, NCK5AI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 2 precursor (CDK5 activator 2) (Cyclin-dependent kinase 5 regulatory subunit 2) (P39) (P39I). | |||||
|
CD5R1_MOUSE
|
||||||
| θ value | 4.38363e-63 (rank : 3) | NC score | 0.931363 (rank : 3) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P61809, Q62938 | Gene names | Cdk5r1, Cdk5r, Nck5a | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 1 precursor (CDK5 activator 1) (Cyclin-dependent kinase 5 regulatory subunit 1) (Tau protein kinase II 23 kDa subunit) (TPKII regulatory subunit) (p23) (p25) (p35) [Contains: Cyclin-dependent kinase 5 activator 1, p35; Cyclin- dependent kinase 5 activator 1, p25]. | |||||
|
CD5R1_HUMAN
|
||||||
| θ value | 4.10295e-61 (rank : 4) | NC score | 0.929735 (rank : 4) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15078 | Gene names | CDK5R1, CDK5R, NCK5A | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 1 precursor (CDK5 activator 1) (Cyclin-dependent kinase 5 regulatory subunit 1) (Tau protein kinase II 23 kDa subunit) (TPKII regulatory subunit) (p23) (p25) (p35) [Contains: Cyclin-dependent kinase 5 activator 1, p35; Cyclin- dependent kinase 5 activator 1, p25]. | |||||
|
CEL_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 5) | NC score | 0.077341 (rank : 6) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 6) | NC score | 0.054567 (rank : 24) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 7) | NC score | 0.075146 (rank : 7) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.086579 (rank : 5) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.060993 (rank : 16) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.056809 (rank : 20) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
CD2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.065637 (rank : 15) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 12) | NC score | 0.048430 (rank : 32) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 13) | NC score | 0.017806 (rank : 54) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
SON_MOUSE
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.057911 (rank : 18) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.016045 (rank : 55) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
IRS2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.058423 (rank : 17) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.038405 (rank : 38) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.066207 (rank : 14) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
MISS_MOUSE
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.048453 (rank : 31) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
PITM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.019580 (rank : 53) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
INVS_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.010708 (rank : 61) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.025577 (rank : 48) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MARK2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.004287 (rank : 70) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q05512, Q6PDR4, Q8BR95 | Gene names | Mark2, Emk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL Motif Kinase) (EMK1). | |||||
|
SCAP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.029650 (rank : 43) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12770, Q8N2E0, Q8WUA1 | Gene names | SCAP, KIAA0199 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein cleavage-activating protein (SREBP cleavage-activating protein) (SCAP). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.049826 (rank : 30) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.044456 (rank : 33) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
FXL19_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.029094 (rank : 45) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PCT2, Q8N789, Q9NT14 | Gene names | FBXL19, FBL19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
FXL19_MOUSE
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.029108 (rank : 44) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6PB97, Q7TSH0, Q8BIB4 | Gene names | Fbxl19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
IDD_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.021104 (rank : 50) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98154, Q61844 | Gene names | Dgcr2, Dgsc, Idd, Sez-12, Sez12 | |||
|
Domain Architecture |
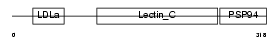
|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor (Seizure-related membrane-bound adhesion protein). | |||||
|
NFAC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.020121 (rank : 52) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
TACC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.020217 (rank : 51) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
CIC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.040805 (rank : 36) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
MSX1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.006282 (rank : 68) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28360, Q96NY4 | Gene names | MSX1, HOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MSX-1 (Msh homeobox 1-like protein) (Hox-7). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.030068 (rank : 41) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PP16A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.012959 (rank : 59) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96I34 | Gene names | PPP1R16A, MYPT3 | |||
|
Domain Architecture |
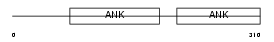
|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase- targeting subunit 3). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.025890 (rank : 47) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.039495 (rank : 37) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
GAK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.007308 (rank : 67) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.022552 (rank : 49) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
PAIP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.034198 (rank : 39) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H074, O60455, Q96B61, Q9BS63 | Gene names | PAIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein-interacting protein 1 (Poly(A)-binding protein-interacting protein 1) (PABP-interacting protein 1) (PAIP-1). | |||||
|
ZN516_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.010549 (rank : 62) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
CTDP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.030034 (rank : 42) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.028572 (rank : 46) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
LIMD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.013087 (rank : 58) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UGP4, Q9BQQ9, Q9NQ47 | Gene names | LIMD1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
SON_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.041119 (rank : 35) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.003888 (rank : 71) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
ADA19_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.007659 (rank : 66) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H013, Q9BZL5, Q9UHP2 | Gene names | ADAM19, MLTNB | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta) (Metalloprotease and disintegrin dentritic antigen marker) (MADDAM). | |||||
|
AKAP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.013434 (rank : 57) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.066281 (rank : 13) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DAF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.008806 (rank : 63) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61475, P97732, Q61397 | Gene names | Cd55a, Daf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor, GPI-anchored precursor (DAF-GPI) (CD55 antigen). | |||||
|
HGFA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.005827 (rank : 69) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04756, Q14726 | Gene names | HGFAC | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.007958 (rank : 64) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.032632 (rank : 40) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PRRT3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.042911 (rank : 34) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SAM14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.013502 (rank : 56) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZD0, Q8N2X0 | Gene names | SAMD14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 14. | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.007889 (rank : 65) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.012092 (rank : 60) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.051811 (rank : 26) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.053595 (rank : 25) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CT166_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.070890 (rank : 8) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H1L0 | Gene names | C20orf166 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf166. | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.056459 (rank : 21) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.069447 (rank : 9) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.066795 (rank : 12) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.069012 (rank : 10) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.068451 (rank : 11) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.054875 (rank : 23) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
PRPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.057318 (rank : 19) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PRPE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.054922 (rank : 22) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.051567 (rank : 27) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.051320 (rank : 28) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.050382 (rank : 29) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
CD5R2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.52337e-140 (rank : 1) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O35926, O35277 | Gene names | Cdk5r2, Nck5ai | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 2 precursor (CDK5 activator 2) (Cyclin-dependent kinase 5 regulatory subunit 2) (P39) (P39I). | |||||
|
CD5R2_HUMAN
|
||||||
| NC score | 0.985870 (rank : 2) | θ value | 2.51683e-135 (rank : 2) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13319, Q4ZFW6 | Gene names | CDK5R2, NCK5AI | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 2 precursor (CDK5 activator 2) (Cyclin-dependent kinase 5 regulatory subunit 2) (P39) (P39I). | |||||
|
CD5R1_MOUSE
|
||||||
| NC score | 0.931363 (rank : 3) | θ value | 4.38363e-63 (rank : 3) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P61809, Q62938 | Gene names | Cdk5r1, Cdk5r, Nck5a | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 1 precursor (CDK5 activator 1) (Cyclin-dependent kinase 5 regulatory subunit 1) (Tau protein kinase II 23 kDa subunit) (TPKII regulatory subunit) (p23) (p25) (p35) [Contains: Cyclin-dependent kinase 5 activator 1, p35; Cyclin- dependent kinase 5 activator 1, p25]. | |||||
|
CD5R1_HUMAN
|
||||||
| NC score | 0.929735 (rank : 4) | θ value | 4.10295e-61 (rank : 4) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15078 | Gene names | CDK5R1, CDK5R, NCK5A | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 5 activator 1 precursor (CDK5 activator 1) (Cyclin-dependent kinase 5 regulatory subunit 1) (Tau protein kinase II 23 kDa subunit) (TPKII regulatory subunit) (p23) (p25) (p35) [Contains: Cyclin-dependent kinase 5 activator 1, p35; Cyclin- dependent kinase 5 activator 1, p25]. | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.086579 (rank : 5) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.077341 (rank : 6) | θ value | 4.1701e-05 (rank : 5) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.075146 (rank : 7) | θ value | 0.0330416 (rank : 7) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
CT166_HUMAN
|
||||||
| NC score | 0.070890 (rank : 8) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H1L0 | Gene names | C20orf166 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf166. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.069447 (rank : 9) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.069012 (rank : 10) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.068451 (rank : 11) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.066795 (rank : 12) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.066281 (rank : 13) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.066207 (rank : 14) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
CD2_HUMAN
|
||||||
| NC score | 0.065637 (rank : 15) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.060993 (rank : 16) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
IRS2_MOUSE
|
||||||
| NC score | 0.058423 (rank : 17) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.057911 (rank : 18) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.057318 (rank : 19) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.056809 (rank : 20) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.056459 (rank : 21) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PRPE_HUMAN
|
||||||
| NC score | 0.054922 (rank : 22) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.054875 (rank : 23) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.054567 (rank : 24) | θ value | 0.0193708 (rank : 6) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.053595 (rank : 25) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.051811 (rank : 26) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.051567 (rank : 27) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.051320 (rank : 28) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.050382 (rank : 29) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.049826 (rank : 30) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.048453 (rank : 31) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.048430 (rank : 32) | θ value | 0.62314 (rank : 12) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.044456 (rank : 33) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
PRRT3_MOUSE
|
||||||
| NC score | 0.042911 (rank : 34) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PE13 | Gene names | Prrt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.041119 (rank : 35) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.040805 (rank : 36) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.039495 (rank : 37) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.038405 (rank : 38) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
PAIP1_HUMAN
|
||||||
| NC score | 0.034198 (rank : 39) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H074, O60455, Q96B61, Q9BS63 | Gene names | PAIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein-interacting protein 1 (Poly(A)-binding protein-interacting protein 1) (PABP-interacting protein 1) (PAIP-1). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.032632 (rank : 40) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.030068 (rank : 41) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
CTDP1_HUMAN
|
||||||
| NC score | 0.030034 (rank : 42) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
SCAP_HUMAN
|
||||||
| NC score | 0.029650 (rank : 43) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12770, Q8N2E0, Q8WUA1 | Gene names | SCAP, KIAA0199 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein cleavage-activating protein (SREBP cleavage-activating protein) (SCAP). | |||||
|
FXL19_MOUSE
|
||||||
| NC score | 0.029108 (rank : 44) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6PB97, Q7TSH0, Q8BIB4 | Gene names | Fbxl19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
FXL19_HUMAN
|
||||||
| NC score | 0.029094 (rank : 45) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PCT2, Q8N789, Q9NT14 | Gene names | FBXL19, FBL19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box/LRR-repeat protein 19 (F-box and leucine-rich repeat protein 19). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.028572 (rank : 46) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.025890 (rank : 47) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.025577 (rank : 48) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.022552 (rank : 49) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
IDD_MOUSE
|
||||||
| NC score | 0.021104 (rank : 50) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98154, Q61844 | Gene names | Dgcr2, Dgsc, Idd, Sez-12, Sez12 | |||
|
Domain Architecture |

|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor (Seizure-related membrane-bound adhesion protein). | |||||
|
TACC2_MOUSE
|
||||||
| NC score | 0.020217 (rank : 51) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
NFAC1_HUMAN
|
||||||
| NC score | 0.020121 (rank : 52) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
PITM1_HUMAN
|
||||||
| NC score | 0.019580 (rank : 53) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.017806 (rank : 54) | θ value | 0.813845 (rank : 13) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.016045 (rank : 55) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
SAM14_HUMAN
|
||||||
| NC score | 0.013502 (rank : 56) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZD0, Q8N2X0 | Gene names | SAMD14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 14. | |||||
|
AKAP1_MOUSE
|
||||||
| NC score | 0.013434 (rank : 57) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
LIMD1_HUMAN
|
||||||
| NC score | 0.013087 (rank : 58) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UGP4, Q9BQQ9, Q9NQ47 | Gene names | LIMD1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
PP16A_HUMAN
|
||||||
| NC score | 0.012959 (rank : 59) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96I34 | Gene names | PPP1R16A, MYPT3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase- targeting subunit 3). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.012092 (rank : 60) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
INVS_HUMAN
|
||||||
| NC score | 0.010708 (rank : 61) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
ZN516_HUMAN
|
||||||
| NC score | 0.010549 (rank : 62) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
DAF1_MOUSE
|
||||||
| NC score | 0.008806 (rank : 63) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61475, P97732, Q61397 | Gene names | Cd55a, Daf1 | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor, GPI-anchored precursor (DAF-GPI) (CD55 antigen). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.007958 (rank : 64) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.007889 (rank : 65) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ADA19_HUMAN
|
||||||
| NC score | 0.007659 (rank : 66) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H013, Q9BZL5, Q9UHP2 | Gene names | ADAM19, MLTNB | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta) (Metalloprotease and disintegrin dentritic antigen marker) (MADDAM). | |||||
|
GAK_HUMAN
|
||||||
| NC score | 0.007308 (rank : 67) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1028 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14976, Q9BVY6 | Gene names | GAK | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
MSX1_HUMAN
|
||||||
| NC score | 0.006282 (rank : 68) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28360, Q96NY4 | Gene names | MSX1, HOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MSX-1 (Msh homeobox 1-like protein) (Hox-7). | |||||
|
HGFA_HUMAN
|
||||||
| NC score | 0.005827 (rank : 69) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04756, Q14726 | Gene names | HGFAC | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
MARK2_MOUSE
|
||||||
| NC score | 0.004287 (rank : 70) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q05512, Q6PDR4, Q8BR95 | Gene names | Mark2, Emk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL Motif Kinase) (EMK1). | |||||
|
ULK1_MOUSE
|
||||||
| NC score | 0.003888 (rank : 71) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||