Please be patient as the page loads
|
BGLR_HUMAN
|
||||||
| SwissProt Accessions | P08236, Q96CL9 | Gene names | GUSB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-glucuronidase precursor (EC 3.2.1.31) (Beta-G1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
BGLR_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08236, Q96CL9 | Gene names | GUSB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-glucuronidase precursor (EC 3.2.1.31) (Beta-G1). | |||||
|
BGLR_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993864 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P12265, Q61601, Q64473, Q64474 | Gene names | Gusb, Gus, Gus-s | |||
|
Domain Architecture |

|
|||||
| Description | Beta-glucuronidase precursor (EC 3.2.1.31). | |||||
|
GUSL1_HUMAN
|
||||||
| θ value | 1.29031e-22 (rank : 3) | NC score | 0.725599 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NQQ0 | Gene names | GUSBL1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-glucuronidase-like protein 1 (SMA3-like protein). | |||||
|
GUSL2_HUMAN
|
||||||
| θ value | 1.29031e-22 (rank : 4) | NC score | 0.725181 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15486, Q969T8, Q9BUH2 | Gene names | SMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-glucuronidase-like protein SMA3. | |||||
|
GUSL3_HUMAN
|
||||||
| θ value | 1.29031e-22 (rank : 5) | NC score | 0.725419 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15487 | Gene names | SMA4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-glucuronidase-like protein SMA4. | |||||
|
GUSL4_HUMAN
|
||||||
| θ value | 1.29031e-22 (rank : 6) | NC score | 0.725778 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15488 | Gene names | SMA5 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-glucuronidase-like protein SMA5. | |||||
|
MANBA_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 7) | NC score | 0.211293 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00462, Q96BC3, Q9NYX9 | Gene names | MANBA, MANB1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-mannosidase precursor (EC 3.2.1.25) (Lysosomal beta A mannosidase) (Mannanase) (Mannase). | |||||
|
MANBA_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 8) | NC score | 0.180466 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2I4, Q3TDB3, Q3TLS7, Q3UQT5, Q99MS1, Q9CRH3 | Gene names | Manba, Bmn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-mannosidase precursor (EC 3.2.1.25) (Lysosomal beta A mannosidase) (Mannanase) (Mannase). | |||||
|
CP238_MOUSE
|
||||||
| θ value | 4.03905 (rank : 9) | NC score | 0.005098 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56655 | Gene names | Cyp2c38 | |||
|
Domain Architecture |
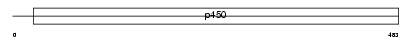
|
|||||
| Description | Cytochrome P450 2C38 (EC 1.14.14.1) (CYPIIC38). | |||||
|
BIR1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 10) | NC score | 0.007765 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUK4, O09124, Q9R030 | Gene names | Birc1b, Naip-rs6, Naip2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1b (Neuronal apoptosis inhibitory protein 2). | |||||
|
PUR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 11) | NC score | 0.022138 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06203 | Gene names | PPAT, GPAT | |||
|
Domain Architecture |

|
|||||
| Description | Amidophosphoribosyltransferase precursor (EC 2.4.2.14) (Glutamine phosphoribosylpyrophosphate amidotransferase) (ATASE) (GPAT). | |||||
|
PEX14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 12) | NC score | 0.015199 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75381, Q8WX51 | Gene names | PEX14 | |||
|
Domain Architecture |
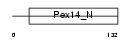
|
|||||
| Description | Peroxisomal membrane protein PEX14 (Peroxin-14) (Peroxisomal membrane anchor protein PEX14) (PTS1 receptor docking protein). | |||||
|
BGLR_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08236, Q96CL9 | Gene names | GUSB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-glucuronidase precursor (EC 3.2.1.31) (Beta-G1). | |||||
|
BGLR_MOUSE
|
||||||
| NC score | 0.993864 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P12265, Q61601, Q64473, Q64474 | Gene names | Gusb, Gus, Gus-s | |||
|
Domain Architecture |

|
|||||
| Description | Beta-glucuronidase precursor (EC 3.2.1.31). | |||||
|
GUSL4_HUMAN
|
||||||
| NC score | 0.725778 (rank : 3) | θ value | 1.29031e-22 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15488 | Gene names | SMA5 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-glucuronidase-like protein SMA5. | |||||
|
GUSL1_HUMAN
|
||||||
| NC score | 0.725599 (rank : 4) | θ value | 1.29031e-22 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NQQ0 | Gene names | GUSBL1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-glucuronidase-like protein 1 (SMA3-like protein). | |||||
|
GUSL3_HUMAN
|
||||||
| NC score | 0.725419 (rank : 5) | θ value | 1.29031e-22 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15487 | Gene names | SMA4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-glucuronidase-like protein SMA4. | |||||
|
GUSL2_HUMAN
|
||||||
| NC score | 0.725181 (rank : 6) | θ value | 1.29031e-22 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15486, Q969T8, Q9BUH2 | Gene names | SMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-glucuronidase-like protein SMA3. | |||||
|
MANBA_HUMAN
|
||||||
| NC score | 0.211293 (rank : 7) | θ value | 1.43324e-05 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00462, Q96BC3, Q9NYX9 | Gene names | MANBA, MANB1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-mannosidase precursor (EC 3.2.1.25) (Lysosomal beta A mannosidase) (Mannanase) (Mannase). | |||||
|
MANBA_MOUSE
|
||||||
| NC score | 0.180466 (rank : 8) | θ value | 0.0113563 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2I4, Q3TDB3, Q3TLS7, Q3UQT5, Q99MS1, Q9CRH3 | Gene names | Manba, Bmn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-mannosidase precursor (EC 3.2.1.25) (Lysosomal beta A mannosidase) (Mannanase) (Mannase). | |||||
|
PUR1_HUMAN
|
||||||
| NC score | 0.022138 (rank : 9) | θ value | 6.88961 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06203 | Gene names | PPAT, GPAT | |||
|
Domain Architecture |

|
|||||
| Description | Amidophosphoribosyltransferase precursor (EC 2.4.2.14) (Glutamine phosphoribosylpyrophosphate amidotransferase) (ATASE) (GPAT). | |||||
|
PEX14_HUMAN
|
||||||
| NC score | 0.015199 (rank : 10) | θ value | 8.99809 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75381, Q8WX51 | Gene names | PEX14 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal membrane protein PEX14 (Peroxin-14) (Peroxisomal membrane anchor protein PEX14) (PTS1 receptor docking protein). | |||||
|
BIR1B_MOUSE
|
||||||
| NC score | 0.007765 (rank : 11) | θ value | 6.88961 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUK4, O09124, Q9R030 | Gene names | Birc1b, Naip-rs6, Naip2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1b (Neuronal apoptosis inhibitory protein 2). | |||||
|
CP238_MOUSE
|
||||||
| NC score | 0.005098 (rank : 12) | θ value | 4.03905 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56655 | Gene names | Cyp2c38 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 2C38 (EC 1.14.14.1) (CYPIIC38). | |||||