Please be patient as the page loads
|
MANBA_MOUSE
|
||||||
| SwissProt Accessions | Q8K2I4, Q3TDB3, Q3TLS7, Q3UQT5, Q99MS1, Q9CRH3 | Gene names | Manba, Bmn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-mannosidase precursor (EC 3.2.1.25) (Lysosomal beta A mannosidase) (Mannanase) (Mannase). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MANBA_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.995767 (rank : 2) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00462, Q96BC3, Q9NYX9 | Gene names | MANBA, MANB1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-mannosidase precursor (EC 3.2.1.25) (Lysosomal beta A mannosidase) (Mannanase) (Mannase). | |||||
|
MANBA_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K2I4, Q3TDB3, Q3TLS7, Q3UQT5, Q99MS1, Q9CRH3 | Gene names | Manba, Bmn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-mannosidase precursor (EC 3.2.1.25) (Lysosomal beta A mannosidase) (Mannanase) (Mannase). | |||||
|
BGLR_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 3) | NC score | 0.168485 (rank : 4) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12265, Q61601, Q64473, Q64474 | Gene names | Gusb, Gus, Gus-s | |||
|
Domain Architecture |

|
|||||
| Description | Beta-glucuronidase precursor (EC 3.2.1.31). | |||||
|
BGLR_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 4) | NC score | 0.180466 (rank : 3) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08236, Q96CL9 | Gene names | GUSB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-glucuronidase precursor (EC 3.2.1.31) (Beta-G1). | |||||
|
LCTL_MOUSE
|
||||||
| θ value | 1.06291 (rank : 5) | NC score | 0.025833 (rank : 6) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K1F9, Q8BPR1, Q8K2M9 | Gene names | Lctl, Klph | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lactase-like protein precursor (Klotho/lactase-phlorizin hydrolase- related protein). | |||||
|
DHPR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 6) | NC score | 0.051472 (rank : 5) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVI4, Q3TT09, Q9D0K4 | Gene names | Qdpr, Dhpr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydropteridine reductase (EC 1.5.1.34) (HDHPR) (Quinoid dihydropteridine reductase). | |||||
|
1A01_HUMAN
|
||||||
| θ value | 3.0926 (rank : 7) | NC score | 0.006043 (rank : 12) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30443, O77964, O78171, Q9MYA3, Q9TP25, Q9TQP5 | Gene names | HLA-A, HLAA | |||
|
Domain Architecture |
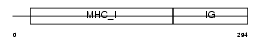
|
|||||
| Description | HLA class I histocompatibility antigen, A-1 alpha chain precursor (MHC class I antigen A*1). | |||||
|
1A36_HUMAN
|
||||||
| θ value | 3.0926 (rank : 8) | NC score | 0.006011 (rank : 13) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30455, Q9MY89 | Gene names | HLA-A, HLAA | |||
|
Domain Architecture |
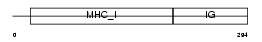
|
|||||
| Description | HLA class I histocompatibility antigen, A-36 alpha chain precursor (MHC class I antigen A*36) (Aw-36). | |||||
|
ICK_HUMAN
|
||||||
| θ value | 4.03905 (rank : 9) | NC score | 0.002250 (rank : 14) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPZ9, O75985, Q5THL2, Q8IYH8, Q9BX17, Q9NYX3 | Gene names | ICK, KIAA0936 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ICK (EC 2.7.11.22) (Intestinal cell kinase) (hICK) (MAK-related kinase) (MRK) (Laryngeal cancer kinase 2) (LCK2). | |||||
|
DNMBP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 10) | NC score | 0.008789 (rank : 9) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6TXD4, Q6ZQ05, Q8CEW8, Q8R0Y2 | Gene names | Dnmbp, Kiaa1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
TLR10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 11) | NC score | 0.007291 (rank : 10) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXR5, Q5FWG4, Q6UXL3 | Gene names | TLR10 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 10 precursor. | |||||
|
NDST4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 12) | NC score | 0.006283 (rank : 11) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H3R1 | Gene names | NDST4, HSST4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 4 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 4) (NDST- 4) (N-heparan sulfate sulfotransferase 4) (N-HSST 4) [Includes: Heparan sulfate N-deacetylase 4 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 4 (EC 2.8.2.-)]. | |||||
|
PPAL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 13) | NC score | 0.012968 (rank : 8) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11117, Q9BTU7 | Gene names | ACP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal acid phosphatase precursor (EC 3.1.3.2) (LAP). | |||||
|
TCLB4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 14) | NC score | 0.015579 (rank : 7) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56844 | Gene names | Tcl1b4 | |||
|
Domain Architecture |
|
|||||
| Description | TCL1B4 protein. | |||||
|
MANBA_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K2I4, Q3TDB3, Q3TLS7, Q3UQT5, Q99MS1, Q9CRH3 | Gene names | Manba, Bmn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-mannosidase precursor (EC 3.2.1.25) (Lysosomal beta A mannosidase) (Mannanase) (Mannase). | |||||
|
MANBA_HUMAN
|
||||||
| NC score | 0.995767 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00462, Q96BC3, Q9NYX9 | Gene names | MANBA, MANB1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-mannosidase precursor (EC 3.2.1.25) (Lysosomal beta A mannosidase) (Mannanase) (Mannase). | |||||
|
BGLR_HUMAN
|
||||||
| NC score | 0.180466 (rank : 3) | θ value | 0.0113563 (rank : 4) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08236, Q96CL9 | Gene names | GUSB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-glucuronidase precursor (EC 3.2.1.31) (Beta-G1). | |||||
|
BGLR_MOUSE
|
||||||
| NC score | 0.168485 (rank : 4) | θ value | 0.00869519 (rank : 3) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12265, Q61601, Q64473, Q64474 | Gene names | Gusb, Gus, Gus-s | |||
|
Domain Architecture |

|
|||||
| Description | Beta-glucuronidase precursor (EC 3.2.1.31). | |||||
|
DHPR_MOUSE
|
||||||
| NC score | 0.051472 (rank : 5) | θ value | 2.36792 (rank : 6) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVI4, Q3TT09, Q9D0K4 | Gene names | Qdpr, Dhpr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydropteridine reductase (EC 1.5.1.34) (HDHPR) (Quinoid dihydropteridine reductase). | |||||
|
LCTL_MOUSE
|
||||||
| NC score | 0.025833 (rank : 6) | θ value | 1.06291 (rank : 5) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K1F9, Q8BPR1, Q8K2M9 | Gene names | Lctl, Klph | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lactase-like protein precursor (Klotho/lactase-phlorizin hydrolase- related protein). | |||||
|
TCLB4_MOUSE
|
||||||
| NC score | 0.015579 (rank : 7) | θ value | 8.99809 (rank : 14) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56844 | Gene names | Tcl1b4 | |||
|
Domain Architecture |

|
|||||
| Description | TCL1B4 protein. | |||||
|
PPAL_HUMAN
|
||||||
| NC score | 0.012968 (rank : 8) | θ value | 8.99809 (rank : 13) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11117, Q9BTU7 | Gene names | ACP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal acid phosphatase precursor (EC 3.1.3.2) (LAP). | |||||
|
DNMBP_MOUSE
|
||||||
| NC score | 0.008789 (rank : 9) | θ value | 6.88961 (rank : 10) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6TXD4, Q6ZQ05, Q8CEW8, Q8R0Y2 | Gene names | Dnmbp, Kiaa1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
TLR10_HUMAN
|
||||||
| NC score | 0.007291 (rank : 10) | θ value | 6.88961 (rank : 11) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXR5, Q5FWG4, Q6UXL3 | Gene names | TLR10 | |||
|
Domain Architecture |

|
|||||
| Description | Toll-like receptor 10 precursor. | |||||
|
NDST4_HUMAN
|
||||||
| NC score | 0.006283 (rank : 11) | θ value | 8.99809 (rank : 12) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H3R1 | Gene names | NDST4, HSST4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 4 (EC 2.8.2.8) (Glucosaminyl N-deacetylase/N-sulfotransferase 4) (NDST- 4) (N-heparan sulfate sulfotransferase 4) (N-HSST 4) [Includes: Heparan sulfate N-deacetylase 4 (EC 3.-.-.-); Heparan sulfate N- sulfotransferase 4 (EC 2.8.2.-)]. | |||||
|
1A01_HUMAN
|
||||||
| NC score | 0.006043 (rank : 12) | θ value | 3.0926 (rank : 7) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30443, O77964, O78171, Q9MYA3, Q9TP25, Q9TQP5 | Gene names | HLA-A, HLAA | |||
|
Domain Architecture |

|
|||||
| Description | HLA class I histocompatibility antigen, A-1 alpha chain precursor (MHC class I antigen A*1). | |||||
|
1A36_HUMAN
|
||||||
| NC score | 0.006011 (rank : 13) | θ value | 3.0926 (rank : 8) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30455, Q9MY89 | Gene names | HLA-A, HLAA | |||
|
Domain Architecture |

|
|||||
| Description | HLA class I histocompatibility antigen, A-36 alpha chain precursor (MHC class I antigen A*36) (Aw-36). | |||||
|
ICK_HUMAN
|
||||||
| NC score | 0.002250 (rank : 14) | θ value | 4.03905 (rank : 9) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPZ9, O75985, Q5THL2, Q8IYH8, Q9BX17, Q9NYX3 | Gene names | ICK, KIAA0936 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ICK (EC 2.7.11.22) (Intestinal cell kinase) (hICK) (MAK-related kinase) (MRK) (Laryngeal cancer kinase 2) (LCK2). | |||||