Please be patient as the page loads
|
ASCC2_HUMAN
|
||||||
| SwissProt Accessions | Q9H1I8, Q4TT54, Q8TAZ0, Q9H711, Q9H9D6 | Gene names | ASCC2, ASC1P100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 2 (ASC-1 complex subunit p100) (Trip4 complex subunit p100). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ASCC2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H1I8, Q4TT54, Q8TAZ0, Q9H711, Q9H9D6 | Gene names | ASCC2, ASC1P100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 2 (ASC-1 complex subunit p100) (Trip4 complex subunit p100). | |||||
|
ASCC2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989382 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WR3, Q8BKM9, Q8BX60, Q8R1B9 | Gene names | Ascc2, Asc1p100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 2 (ASC-1 complex subunit p100) (Trip4 complex subunit p100). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 3) | NC score | 0.056817 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 4) | NC score | 0.050106 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
KIF1B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 5) | NC score | 0.026310 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60333, Q96Q94, Q9BV80, Q9P280 | Gene names | KIF1B, KIAA0591, KIAA1448 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1B (Klp). | |||||
|
CMGA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 6) | NC score | 0.045583 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
KIF1B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 7) | NC score | 0.023239 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60575, Q9R0B4, Q9WVE5, Q9Z119 | Gene names | Kif1b | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1B. | |||||
|
LAMC1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 8) | NC score | 0.011540 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P02468 | Gene names | Lamc1, Lamb-2, Lamc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
KCNN2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 9) | NC score | 0.020576 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
ECHP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 10) | NC score | 0.020779 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08426 | Gene names | EHHADH, ECHD | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal bifunctional enzyme (PBE) (PBFE) [Includes: Enoyl-CoA hydratase (EC 4.2.1.17); 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8); 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35)]. | |||||
|
MAGA8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 11) | NC score | 0.014554 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43361 | Gene names | MAGEA8, MAGE8 | |||
|
Domain Architecture |
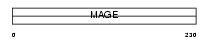
|
|||||
| Description | Melanoma-associated antigen 8 (MAGE-8 antigen). | |||||
|
CA082_HUMAN
|
||||||
| θ value | 5.27518 (rank : 12) | NC score | 0.022766 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IXW5, Q49AS7, Q9H8Y2 | Gene names | C1orf82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0385 protein C1orf82. | |||||
|
CROP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 13) | NC score | 0.014526 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
FETA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 14) | NC score | 0.019756 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02772, Q3UJD0 | Gene names | Afp | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein precursor (Alpha-fetoglobulin) (Alpha-1- fetoprotein). | |||||
|
DOK4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 15) | NC score | 0.012755 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99KE3, Q3U0X3 | Gene names | Dok4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 4 (Downstream of tyrosine kinase 4). | |||||
|
SIN3B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 16) | NC score | 0.012697 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75182, Q68GC2, Q6P4B8, Q8TB34, Q9BSC8 | Gene names | SIN3B, KIAA0700 | |||
|
Domain Architecture |
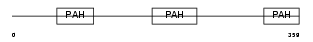
|
|||||
| Description | Paired amphipathic helix protein Sin3b (Transcriptional corepressor Sin3b) (Histone deacetylase complex subunit Sin3b). | |||||
|
TULP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 17) | NC score | 0.011567 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z273 | Gene names | Tulp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
VEZA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.023223 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBM0, Q9H2F4, Q9H2U5, Q9NT70, Q9NVW0, Q9UF91 | Gene names | VEZT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vezatin. | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 19) | NC score | 0.005139 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
MAGB6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.010150 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
NEBU_HUMAN
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.009407 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20929, Q15346 | Gene names | NEB | |||
|
Domain Architecture |
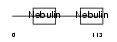
|
|||||
| Description | Nebulin. | |||||
|
RN165_HUMAN
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.012586 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZSG1 | Gene names | RNF165 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 165. | |||||
|
TPSNR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.008940 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VD31 | Gene names | Tapbpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tapasin-related protein precursor (TAPASIN-R) (Tapasin-like) (TAP- binding protein-related protein) (TAPBP-R) (TAP-binding protein-like). | |||||
|
WDHD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.008343 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75717 | Gene names | WDHD1, AND1 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
ASCC2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H1I8, Q4TT54, Q8TAZ0, Q9H711, Q9H9D6 | Gene names | ASCC2, ASC1P100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 2 (ASC-1 complex subunit p100) (Trip4 complex subunit p100). | |||||
|
ASCC2_MOUSE
|
||||||
| NC score | 0.989382 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WR3, Q8BKM9, Q8BX60, Q8R1B9 | Gene names | Ascc2, Asc1p100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 2 (ASC-1 complex subunit p100) (Trip4 complex subunit p100). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.056817 (rank : 3) | θ value | 0.0330416 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.050106 (rank : 4) | θ value | 0.125558 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CMGA_HUMAN
|
||||||
| NC score | 0.045583 (rank : 5) | θ value | 0.62314 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
KIF1B_HUMAN
|
||||||
| NC score | 0.026310 (rank : 6) | θ value | 0.47712 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60333, Q96Q94, Q9BV80, Q9P280 | Gene names | KIF1B, KIAA0591, KIAA1448 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1B (Klp). | |||||
|
KIF1B_MOUSE
|
||||||
| NC score | 0.023239 (rank : 7) | θ value | 0.813845 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60575, Q9R0B4, Q9WVE5, Q9Z119 | Gene names | Kif1b | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1B. | |||||
|
VEZA_HUMAN
|
||||||
| NC score | 0.023223 (rank : 8) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBM0, Q9H2F4, Q9H2U5, Q9NT70, Q9NVW0, Q9UF91 | Gene names | VEZT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vezatin. | |||||
|
CA082_HUMAN
|
||||||
| NC score | 0.022766 (rank : 9) | θ value | 5.27518 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IXW5, Q49AS7, Q9H8Y2 | Gene names | C1orf82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0385 protein C1orf82. | |||||
|
ECHP_HUMAN
|
||||||
| NC score | 0.020779 (rank : 10) | θ value | 3.0926 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08426 | Gene names | EHHADH, ECHD | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal bifunctional enzyme (PBE) (PBFE) [Includes: Enoyl-CoA hydratase (EC 4.2.1.17); 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8); 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35)]. | |||||
|
KCNN2_MOUSE
|
||||||
| NC score | 0.020576 (rank : 11) | θ value | 1.38821 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
FETA_MOUSE
|
||||||
| NC score | 0.019756 (rank : 12) | θ value | 5.27518 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02772, Q3UJD0 | Gene names | Afp | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein precursor (Alpha-fetoglobulin) (Alpha-1- fetoprotein). | |||||
|
MAGA8_HUMAN
|
||||||
| NC score | 0.014554 (rank : 13) | θ value | 3.0926 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43361 | Gene names | MAGEA8, MAGE8 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 8 (MAGE-8 antigen). | |||||
|
CROP_HUMAN
|
||||||
| NC score | 0.014526 (rank : 14) | θ value | 5.27518 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
DOK4_MOUSE
|
||||||
| NC score | 0.012755 (rank : 15) | θ value | 6.88961 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99KE3, Q3U0X3 | Gene names | Dok4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Docking protein 4 (Downstream of tyrosine kinase 4). | |||||
|
SIN3B_HUMAN
|
||||||
| NC score | 0.012697 (rank : 16) | θ value | 6.88961 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75182, Q68GC2, Q6P4B8, Q8TB34, Q9BSC8 | Gene names | SIN3B, KIAA0700 | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3b (Transcriptional corepressor Sin3b) (Histone deacetylase complex subunit Sin3b). | |||||
|
RN165_HUMAN
|
||||||
| NC score | 0.012586 (rank : 17) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZSG1 | Gene names | RNF165 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 165. | |||||
|
TULP1_MOUSE
|
||||||
| NC score | 0.011567 (rank : 18) | θ value | 6.88961 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z273 | Gene names | Tulp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
LAMC1_MOUSE
|
||||||
| NC score | 0.011540 (rank : 19) | θ value | 0.813845 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P02468 | Gene names | Lamc1, Lamb-2, Lamc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
MAGB6_HUMAN
|
||||||
| NC score | 0.010150 (rank : 20) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N7X4, Q6GS19, Q9H219 | Gene names | MAGEB6 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B6 (MAGE-B6 antigen). | |||||
|
NEBU_HUMAN
|
||||||
| NC score | 0.009407 (rank : 21) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20929, Q15346 | Gene names | NEB | |||
|
Domain Architecture |

|
|||||
| Description | Nebulin. | |||||
|
TPSNR_MOUSE
|
||||||
| NC score | 0.008940 (rank : 22) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VD31 | Gene names | Tapbpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tapasin-related protein precursor (TAPASIN-R) (Tapasin-like) (TAP- binding protein-related protein) (TAPBP-R) (TAP-binding protein-like). | |||||
|
WDHD1_HUMAN
|
||||||
| NC score | 0.008343 (rank : 23) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75717 | Gene names | WDHD1, AND1 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.005139 (rank : 24) | θ value | 8.99809 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||