Please be patient as the page loads
|
AMPD3_HUMAN
|
||||||
| SwissProt Accessions | Q01432 | Gene names | AMPD3 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase isoform E) (Erythrocyte AMP deaminase). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AMPD1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.990859 (rank : 3) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23109 | Gene names | AMPD1 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 1 (EC 3.5.4.6) (Myoadenylate deaminase) (AMP deaminase isoform M). | |||||
|
AMPD2_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.979338 (rank : 5) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01433, Q14856, Q14857, Q16686, Q16687, Q16688, Q16729, Q96IA1, Q9UDX8, Q9UDX9, Q9UMU4 | Gene names | AMPD2 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 2 (EC 3.5.4.6) (AMP deaminase isoform L). | |||||
|
AMPD2_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.980796 (rank : 4) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBT5, Q91YI2 | Gene names | Ampd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AMP deaminase 2 (EC 3.5.4.6). | |||||
|
AMPD3_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q01432 | Gene names | AMPD3 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase isoform E) (Erythrocyte AMP deaminase). | |||||
|
AMPD3_MOUSE
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.997550 (rank : 2) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08739, O88692 | Gene names | Ampd3 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase isoform E) (AMP deaminase H-type) (Heart-type AMPD). | |||||
|
CECR1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 6) | NC score | 0.270569 (rank : 6) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZK5, Q96K41 | Gene names | CECR1 | |||
|
Domain Architecture |
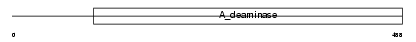
|
|||||
| Description | Cat eye syndrome critical region protein 1 precursor. | |||||
|
NELFA_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 7) | NC score | 0.095691 (rank : 7) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
NELFA_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.090946 (rank : 8) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H3P2, O95392 | Gene names | WHSC2, NELFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 protein). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 9) | NC score | 0.018640 (rank : 14) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 10) | NC score | 0.020383 (rank : 13) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
CAD23_MOUSE
|
||||||
| θ value | 2.36792 (rank : 11) | NC score | 0.008961 (rank : 24) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99PF4, Q99NH1, Q9D4N9 | Gene names | Cdh23 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-23 precursor (Otocadherin). | |||||
|
CD6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 12) | NC score | 0.018481 (rank : 15) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30203, Q9UMF2, Q9Y4K7, Q9Y4K8, Q9Y4K9, Q9Y4L0 | Gene names | CD6 | |||
|
Domain Architecture |
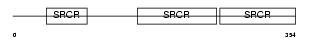
|
|||||
| Description | T-cell differentiation antigen CD6 precursor (T12) (TP120). | |||||
|
PHLA1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 13) | NC score | 0.020842 (rank : 11) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62392, Q3TY48, Q3UG87 | Gene names | Phlda1, Tdag51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Proline- and glutamine-rich protein) (PQR protein). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.016520 (rank : 17) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ABI2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.013427 (rank : 19) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYB9, Q13147, Q13249, Q13801, Q9BV70 | Gene names | ABI2, ARGBPIA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2) (Abl-binding protein 3) (AblBP3) (Arg-binding protein 1) (ArgBP1). | |||||
|
ABR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.012523 (rank : 20) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
AQR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.014893 (rank : 18) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60306, Q2YDX9, Q6IRU8, Q6PIC8 | Gene names | AQR, KIAA0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius (Intron-binding protein of 160 kDa) (IBP160). | |||||
|
IFT81_HUMAN
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.009654 (rank : 23) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
IFT81_MOUSE
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.008712 (rank : 25) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
ABI2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.012415 (rank : 21) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62484, Q6PHU3 | Gene names | Abi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2). | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.005657 (rank : 29) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CAD23_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.007988 (rank : 26) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H251, Q6UWW1, Q9H4K9 | Gene names | CDH23, KIAA1774 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-23 precursor (Otocadherin). | |||||
|
CSMD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.005922 (rank : 28) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
PDGFD_MOUSE
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.023648 (rank : 10) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q925I7, Q3URF6, Q8K2L3, Q9D1L8 | Gene names | Pdgfd, Scdgfb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor chain precursor (PDGF D) (Spinal cord- derived growth factor B) (SCDGF-B). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.002223 (rank : 32) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
ORC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.023926 (rank : 9) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JK30 | Gene names | Orc3l, Orc3 | |||
|
Domain Architecture |
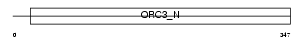
|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
PDE4D_HUMAN
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.004258 (rank : 31) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
RRMJ3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.020775 (rank : 12) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBE9, Q3ULI1, Q921I7 | Gene names | Ftsj3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
ABTB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.005168 (rank : 30) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q969K4, Q6ZNU9, Q71MF1, Q96S62, Q96S63 | Gene names | ABTB1, BPOZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 1 (Elongation factor 1A-binding protein). | |||||
|
CG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.011427 (rank : 22) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.000982 (rank : 33) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
NOP14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.017583 (rank : 16) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
NP1L3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.006557 (rank : 27) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
AMPD3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q01432 | Gene names | AMPD3 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase isoform E) (Erythrocyte AMP deaminase). | |||||
|
AMPD3_MOUSE
|
||||||
| NC score | 0.997550 (rank : 2) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08739, O88692 | Gene names | Ampd3 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 3 (EC 3.5.4.6) (AMP deaminase isoform E) (AMP deaminase H-type) (Heart-type AMPD). | |||||
|
AMPD1_HUMAN
|
||||||
| NC score | 0.990859 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23109 | Gene names | AMPD1 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 1 (EC 3.5.4.6) (Myoadenylate deaminase) (AMP deaminase isoform M). | |||||
|
AMPD2_MOUSE
|
||||||
| NC score | 0.980796 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBT5, Q91YI2 | Gene names | Ampd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AMP deaminase 2 (EC 3.5.4.6). | |||||
|
AMPD2_HUMAN
|
||||||
| NC score | 0.979338 (rank : 5) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01433, Q14856, Q14857, Q16686, Q16687, Q16688, Q16729, Q96IA1, Q9UDX8, Q9UDX9, Q9UMU4 | Gene names | AMPD2 | |||
|
Domain Architecture |

|
|||||
| Description | AMP deaminase 2 (EC 3.5.4.6) (AMP deaminase isoform L). | |||||
|
CECR1_HUMAN
|
||||||
| NC score | 0.270569 (rank : 6) | θ value | 0.000121331 (rank : 6) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZK5, Q96K41 | Gene names | CECR1 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 1 precursor. | |||||
|
NELFA_MOUSE
|
||||||
| NC score | 0.095691 (rank : 7) | θ value | 0.0193708 (rank : 7) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
NELFA_HUMAN
|
||||||
| NC score | 0.090946 (rank : 8) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H3P2, O95392 | Gene names | WHSC2, NELFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 protein). | |||||
|
ORC3_MOUSE
|
||||||
| NC score | 0.023926 (rank : 9) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JK30 | Gene names | Orc3l, Orc3 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 3 (Origin recognition complex subunit Latheo). | |||||
|
PDGFD_MOUSE
|
||||||
| NC score | 0.023648 (rank : 10) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q925I7, Q3URF6, Q8K2L3, Q9D1L8 | Gene names | Pdgfd, Scdgfb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor chain precursor (PDGF D) (Spinal cord- derived growth factor B) (SCDGF-B). | |||||
|
PHLA1_MOUSE
|
||||||
| NC score | 0.020842 (rank : 11) | θ value | 2.36792 (rank : 13) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62392, Q3TY48, Q3UG87 | Gene names | Phlda1, Tdag51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Proline- and glutamine-rich protein) (PQR protein). | |||||
|
RRMJ3_MOUSE
|
||||||
| NC score | 0.020775 (rank : 12) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DBE9, Q3ULI1, Q921I7 | Gene names | Ftsj3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.020383 (rank : 13) | θ value | 1.38821 (rank : 10) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.018640 (rank : 14) | θ value | 0.62314 (rank : 9) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CD6_HUMAN
|
||||||
| NC score | 0.018481 (rank : 15) | θ value | 2.36792 (rank : 12) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30203, Q9UMF2, Q9Y4K7, Q9Y4K8, Q9Y4K9, Q9Y4L0 | Gene names | CD6 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell differentiation antigen CD6 precursor (T12) (TP120). | |||||
|
NOP14_HUMAN
|
||||||
| NC score | 0.017583 (rank : 16) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.016520 (rank : 17) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
AQR_HUMAN
|
||||||
| NC score | 0.014893 (rank : 18) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60306, Q2YDX9, Q6IRU8, Q6PIC8 | Gene names | AQR, KIAA0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius (Intron-binding protein of 160 kDa) (IBP160). | |||||
|
ABI2_HUMAN
|
||||||
| NC score | 0.013427 (rank : 19) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYB9, Q13147, Q13249, Q13801, Q9BV70 | Gene names | ABI2, ARGBPIA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2) (Abl-binding protein 3) (AblBP3) (Arg-binding protein 1) (ArgBP1). | |||||
|
ABR_HUMAN
|
||||||
| NC score | 0.012523 (rank : 20) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12979, Q13693, Q13694 | Gene names | ABR | |||
|
Domain Architecture |

|
|||||
| Description | Active breakpoint cluster region-related protein. | |||||
|
ABI2_MOUSE
|
||||||
| NC score | 0.012415 (rank : 21) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P62484, Q6PHU3 | Gene names | Abi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2). | |||||
|
CG1_HUMAN
|
||||||
| NC score | 0.011427 (rank : 22) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
IFT81_HUMAN
|
||||||
| NC score | 0.009654 (rank : 23) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
CAD23_MOUSE
|
||||||
| NC score | 0.008961 (rank : 24) | θ value | 2.36792 (rank : 11) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99PF4, Q99NH1, Q9D4N9 | Gene names | Cdh23 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-23 precursor (Otocadherin). | |||||
|
IFT81_MOUSE
|
||||||
| NC score | 0.008712 (rank : 25) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
CAD23_HUMAN
|
||||||
| NC score | 0.007988 (rank : 26) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H251, Q6UWW1, Q9H4K9 | Gene names | CDH23, KIAA1774 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-23 precursor (Otocadherin). | |||||
|
NP1L3_MOUSE
|
||||||
| NC score | 0.006557 (rank : 27) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
CSMD2_HUMAN
|
||||||
| NC score | 0.005922 (rank : 28) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.005657 (rank : 29) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
ABTB1_HUMAN
|
||||||
| NC score | 0.005168 (rank : 30) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q969K4, Q6ZNU9, Q71MF1, Q96S62, Q96S63 | Gene names | ABTB1, BPOZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 1 (Elongation factor 1A-binding protein). | |||||
|
PDE4D_HUMAN
|
||||||
| NC score | 0.004258 (rank : 31) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08499, O43433, Q13549, Q13550, Q13551, Q7Z2L8, Q8IV84, Q8IVA9, Q8IVD2, Q8IVD3, Q9HCX7 | Gene names | PDE4D | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-specific 3',5'-cyclic phosphodiesterase 4D (EC 3.1.4.17) (DPDE3) (PDE43). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.002223 (rank : 32) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.000982 (rank : 33) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||