Please be patient as the page loads
|
ZP1_MOUSE
|
||||||
| SwissProt Accessions | Q62005, Q62016 | Gene names | Zp1 | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 1 precursor (Zona pellucida glycoprotein 1) (Zp-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ZP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.986833 (rank : 2) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P60852 | Gene names | ZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zona pellucida sperm-binding protein 1 precursor (Zona pellucida glycoprotein 1) (Zp-1). | |||||
|
ZP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62005, Q62016 | Gene names | Zp1 | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 1 precursor (Zona pellucida glycoprotein 1) (Zp-1). | |||||
|
ZP4_HUMAN
|
||||||
| θ value | 6.71607e-88 (rank : 3) | NC score | 0.927023 (rank : 3) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12836 | Gene names | ZP4, ZPB | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 4 precursor (Zona pellucida sperm-binding protein B). | |||||
|
ZP2_MOUSE
|
||||||
| θ value | 5.57106e-42 (rank : 4) | NC score | 0.831933 (rank : 4) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P20239 | Gene names | Zp2, Zp-2, Zpa | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 2 precursor (Zona pellucida glycoprotein ZP2) (Zona pellucida protein A). | |||||
|
ZP2_HUMAN
|
||||||
| θ value | 8.04462e-41 (rank : 5) | NC score | 0.817206 (rank : 5) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q05996 | Gene names | ZP2, ZPA | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 2 precursor (Zona pellucida glycoprotein ZP2) (Zona pellucida protein A). | |||||
|
CUZD1_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 6) | NC score | 0.346197 (rank : 7) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
CUZD1_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 7) | NC score | 0.345311 (rank : 8) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
TECTA_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 8) | NC score | 0.319919 (rank : 10) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75443 | Gene names | TECTA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
TECTA_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 9) | NC score | 0.317386 (rank : 11) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08523 | Gene names | Tecta | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
DMBT1_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 10) | NC score | 0.211830 (rank : 17) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
GP2_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 11) | NC score | 0.381279 (rank : 6) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D733 | Gene names | Gp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreatic secretory granule membrane major glycoprotein GP2 precursor (Pancreatic zymogen granule membrane protein GP-2). | |||||
|
DMBT1_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 12) | NC score | 0.206555 (rank : 18) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UGM3, Q59EX0, Q5JR26, Q6MZN4, Q96DU4, Q9UGM2, Q9UJ57, Q9UKJ4, Q9Y211, Q9Y4V9 | Gene names | DMBT1, GP340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (Glycoprotein 340) (Gp-340) (Surfactant pulmonary-associated D-binding protein). | |||||
|
ZP3_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 13) | NC score | 0.303756 (rank : 13) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P21754, Q06633 | Gene names | ZP3, ZP3A, ZPC | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 3 precursor (Zona pellucida glycoprotein ZP3) (Zona pellucida protein C) (Sperm receptor) (ZP3A/ZP3B). | |||||
|
UROM_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 14) | NC score | 0.200002 (rank : 19) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91X17, Q3TN64, Q3TP60, Q62285 | Gene names | Umod | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin precursor (Tamm-Horsfall urinary glycoprotein) (THP). | |||||
|
GP2_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 15) | NC score | 0.338761 (rank : 9) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P55259, Q13338, Q9UIF1 | Gene names | GP2 | |||
|
Domain Architecture |
|
|||||
| Description | Pancreatic secretory granule membrane major glycoprotein GP2 precursor (Pancreatic zymogen granule membrane protein GP-2) (ZAP75). | |||||
|
UROL1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 16) | NC score | 0.274577 (rank : 15) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5DID3, Q5DID1, Q5DID2 | Gene names | Umodl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin-like 1 precursor (Olfactorin). | |||||
|
UROL1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 17) | NC score | 0.270141 (rank : 16) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5DID0, Q5DIC9, Q6LA40, Q6LA41, Q8N216 | Gene names | UMODL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin-like 1 precursor (Olfactorin). | |||||
|
UROM_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 18) | NC score | 0.185286 (rank : 20) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P07911, Q540J6, Q6ZS84, Q8IYG0 | Gene names | UMOD | |||
|
Domain Architecture |

|
|||||
| Description | Uromodulin precursor (Tamm-Horsfall urinary glycoprotein) (THP). | |||||
|
ZN295_HUMAN
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.010755 (rank : 42) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULJ3, Q6P4R0 | Gene names | ZNF295, KIAA1227, ZBTB21 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 295 (Zinc finger and BTB domain-containing protein 21). | |||||
|
TECTB_HUMAN
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.304411 (rank : 12) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96PL2 | Gene names | TECTB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-tectorin precursor. | |||||
|
TECTB_MOUSE
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.303457 (rank : 14) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08524 | Gene names | Tectb | |||
|
Domain Architecture |

|
|||||
| Description | Beta-tectorin precursor. | |||||
|
CD5_MOUSE
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.078868 (rank : 24) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13379 | Gene names | Cd5, Ly-1 | |||
|
Domain Architecture |
|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen Ly-1) (Lyt-1). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.026416 (rank : 30) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
IGHA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.018213 (rank : 33) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01876 | Gene names | IGHA1 | |||
|
Domain Architecture |
|
|||||
| Description | Ig alpha-1 chain C region. | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.010938 (rank : 41) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
CAH9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.010598 (rank : 43) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHB5, Q8K1G1, Q8VDE4 | Gene names | Ca9, Car9 | |||
|
Domain Architecture |
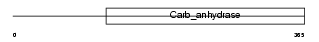
|
|||||
| Description | Carbonic anhydrase 9 precursor (EC 4.2.1.1) (Carbonic anhydrase IX) (Carbonate dehydratase IX) (CA-IX) (CAIX) (Membrane antigen MN homolog). | |||||
|
KCNH2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.014278 (rank : 36) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.014285 (rank : 35) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
XKR5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.019374 (rank : 32) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UX68, Q5GH74 | Gene names | XKR5, XRG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 5. | |||||
|
CD5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.067677 (rank : 25) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P06127 | Gene names | CD5, LEU1 | |||
|
Domain Architecture |
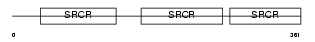
|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen T1/Leu- 1). | |||||
|
KR105_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.011828 (rank : 39) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
PLAC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.024768 (rank : 31) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JI83, Q3UKT4, Q80WW3 | Gene names | Plac1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Placenta-specific protein 1 precursor (EPCS26). | |||||
|
EMX2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.006479 (rank : 46) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04743, Q96NN8, Q9BQF4 | Gene names | EMX2 | |||
|
Domain Architecture |
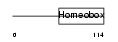
|
|||||
| Description | Homeobox protein EMX2 (Empty spiracles homolog 2) (Empty spiracles- like protein 2). | |||||
|
EMX2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.006465 (rank : 47) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04744, Q80UE7 | Gene names | Emx2, Emx-2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein EMX2 (Empty spiracles homolog 2) (Empty spiracles- like protein 2). | |||||
|
SH3K1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.007946 (rank : 45) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R550, Q8CAL8, Q8CEF6, Q8R545, Q8R546, Q8R547, Q8R548, Q8R549, Q8R551, Q9CTQ9, Q9DC14, Q9JKC3 | Gene names | Sh3kbp1, Ruk, Seta | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (SH3-containing, expressed in tumorigenic astrocytes) (Regulator of ubiquitous kinase) (Ruk). | |||||
|
TSP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.009492 (rank : 44) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35441 | Gene names | Thbs1, Tsp1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.002563 (rank : 50) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.003748 (rank : 48) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.002890 (rank : 49) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
ZN750_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.014309 (rank : 34) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q32MQ0, Q9H899 | Gene names | ZNF750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.013744 (rank : 37) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.013087 (rank : 38) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
IF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.011364 (rank : 40) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27352 | Gene names | GIF, IFMH | |||
|
Domain Architecture |

|
|||||
| Description | Gastric intrinsic factor precursor (Intrinsic factor) (IF) (INF). | |||||
|
C163A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.053912 (rank : 27) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86VB7, Q07898, Q07899, Q07900, Q07901, Q2VLH7 | Gene names | CD163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) (Hemoglobin scavenger receptor) [Contains: Soluble CD163 (sCD163)]. | |||||
|
C163A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.053726 (rank : 28) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q2VLH6, Q2VLH5, Q99MX8 | Gene names | Cd163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) [Contains: Soluble CD163 (sCD163)]. | |||||
|
C163B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.054631 (rank : 26) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NR16, Q6UWC2 | Gene names | CD163L1, CD163B, M160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M160 precursor (CD163 antigen-like 1) (CD163b antigen). | |||||
|
TGBR3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.104573 (rank : 23) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q03167, Q5T2T4, Q5U731, Q9UGI2 | Gene names | TGFBR3 | |||
|
Domain Architecture |

|
|||||
| Description | TGF-beta receptor type III precursor (TGFR-3) (Transforming growth factor beta receptor III) (Betaglycan). | |||||
|
TGBR3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.140786 (rank : 22) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88393, Q6NS72 | Gene names | Tgfbr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta receptor type III precursor (TGFR-3) (Transforming growth factor beta receptor III) (Betaglycan). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.052320 (rank : 29) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.141137 (rank : 21) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10761 | Gene names | Zp3, Zp-3, Zpc | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 3 precursor (Zona pellucida glycoprotein ZP3) (Sperm receptor) (Zona pellucida protein C). | |||||
|
ZP1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62005, Q62016 | Gene names | Zp1 | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 1 precursor (Zona pellucida glycoprotein 1) (Zp-1). | |||||
|
ZP1_HUMAN
|
||||||
| NC score | 0.986833 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P60852 | Gene names | ZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zona pellucida sperm-binding protein 1 precursor (Zona pellucida glycoprotein 1) (Zp-1). | |||||
|
ZP4_HUMAN
|
||||||
| NC score | 0.927023 (rank : 3) | θ value | 6.71607e-88 (rank : 3) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12836 | Gene names | ZP4, ZPB | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 4 precursor (Zona pellucida sperm-binding protein B). | |||||
|
ZP2_MOUSE
|
||||||
| NC score | 0.831933 (rank : 4) | θ value | 5.57106e-42 (rank : 4) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P20239 | Gene names | Zp2, Zp-2, Zpa | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 2 precursor (Zona pellucida glycoprotein ZP2) (Zona pellucida protein A). | |||||
|
ZP2_HUMAN
|
||||||
| NC score | 0.817206 (rank : 5) | θ value | 8.04462e-41 (rank : 5) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q05996 | Gene names | ZP2, ZPA | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 2 precursor (Zona pellucida glycoprotein ZP2) (Zona pellucida protein A). | |||||
|
GP2_MOUSE
|
||||||
| NC score | 0.381279 (rank : 6) | θ value | 3.77169e-06 (rank : 11) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D733 | Gene names | Gp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreatic secretory granule membrane major glycoprotein GP2 precursor (Pancreatic zymogen granule membrane protein GP-2). | |||||
|
CUZD1_HUMAN
|
||||||
| NC score | 0.346197 (rank : 7) | θ value | 6.21693e-09 (rank : 6) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
CUZD1_MOUSE
|
||||||
| NC score | 0.345311 (rank : 8) | θ value | 2.36244e-08 (rank : 7) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
GP2_HUMAN
|
||||||
| NC score | 0.338761 (rank : 9) | θ value | 0.00134147 (rank : 15) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P55259, Q13338, Q9UIF1 | Gene names | GP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic secretory granule membrane major glycoprotein GP2 precursor (Pancreatic zymogen granule membrane protein GP-2) (ZAP75). | |||||
|
TECTA_HUMAN
|
||||||
| NC score | 0.319919 (rank : 10) | θ value | 1.17247e-07 (rank : 8) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75443 | Gene names | TECTA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
TECTA_MOUSE
|
||||||
| NC score | 0.317386 (rank : 11) | θ value | 1.69304e-06 (rank : 9) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08523 | Gene names | Tecta | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
TECTB_HUMAN
|
||||||
| NC score | 0.304411 (rank : 12) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96PL2 | Gene names | TECTB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-tectorin precursor. | |||||
|
ZP3_HUMAN
|
||||||
| NC score | 0.303756 (rank : 13) | θ value | 0.000461057 (rank : 13) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P21754, Q06633 | Gene names | ZP3, ZP3A, ZPC | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 3 precursor (Zona pellucida glycoprotein ZP3) (Zona pellucida protein C) (Sperm receptor) (ZP3A/ZP3B). | |||||
|
TECTB_MOUSE
|
||||||
| NC score | 0.303457 (rank : 14) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08524 | Gene names | Tectb | |||
|
Domain Architecture |

|
|||||
| Description | Beta-tectorin precursor. | |||||
|
UROL1_MOUSE
|
||||||
| NC score | 0.274577 (rank : 15) | θ value | 0.00869519 (rank : 16) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5DID3, Q5DID1, Q5DID2 | Gene names | Umodl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin-like 1 precursor (Olfactorin). | |||||
|
UROL1_HUMAN
|
||||||
| NC score | 0.270141 (rank : 16) | θ value | 0.0148317 (rank : 17) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5DID0, Q5DIC9, Q6LA40, Q6LA41, Q8N216 | Gene names | UMODL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin-like 1 precursor (Olfactorin). | |||||
|
DMBT1_MOUSE
|
||||||
| NC score | 0.211830 (rank : 17) | θ value | 2.21117e-06 (rank : 10) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
DMBT1_HUMAN
|
||||||
| NC score | 0.206555 (rank : 18) | θ value | 1.43324e-05 (rank : 12) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UGM3, Q59EX0, Q5JR26, Q6MZN4, Q96DU4, Q9UGM2, Q9UJ57, Q9UKJ4, Q9Y211, Q9Y4V9 | Gene names | DMBT1, GP340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (Glycoprotein 340) (Gp-340) (Surfactant pulmonary-associated D-binding protein). | |||||
|
UROM_MOUSE
|
||||||
| NC score | 0.200002 (rank : 19) | θ value | 0.00102713 (rank : 14) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91X17, Q3TN64, Q3TP60, Q62285 | Gene names | Umod | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin precursor (Tamm-Horsfall urinary glycoprotein) (THP). | |||||
|
UROM_HUMAN
|
||||||
| NC score | 0.185286 (rank : 20) | θ value | 0.0431538 (rank : 18) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P07911, Q540J6, Q6ZS84, Q8IYG0 | Gene names | UMOD | |||
|
Domain Architecture |

|
|||||
| Description | Uromodulin precursor (Tamm-Horsfall urinary glycoprotein) (THP). | |||||
|
ZP3_MOUSE
|
||||||
| NC score | 0.141137 (rank : 21) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10761 | Gene names | Zp3, Zp-3, Zpc | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 3 precursor (Zona pellucida glycoprotein ZP3) (Sperm receptor) (Zona pellucida protein C). | |||||
|
TGBR3_MOUSE
|
||||||
| NC score | 0.140786 (rank : 22) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88393, Q6NS72 | Gene names | Tgfbr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta receptor type III precursor (TGFR-3) (Transforming growth factor beta receptor III) (Betaglycan). | |||||
|
TGBR3_HUMAN
|
||||||
| NC score | 0.104573 (rank : 23) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q03167, Q5T2T4, Q5U731, Q9UGI2 | Gene names | TGFBR3 | |||
|
Domain Architecture |

|
|||||
| Description | TGF-beta receptor type III precursor (TGFR-3) (Transforming growth factor beta receptor III) (Betaglycan). | |||||
|
CD5_MOUSE
|
||||||
| NC score | 0.078868 (rank : 24) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13379 | Gene names | Cd5, Ly-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen Ly-1) (Lyt-1). | |||||
|
CD5_HUMAN
|
||||||
| NC score | 0.067677 (rank : 25) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P06127 | Gene names | CD5, LEU1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen T1/Leu- 1). | |||||
|
C163B_HUMAN
|
||||||
| NC score | 0.054631 (rank : 26) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NR16, Q6UWC2 | Gene names | CD163L1, CD163B, M160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M160 precursor (CD163 antigen-like 1) (CD163b antigen). | |||||
|
C163A_HUMAN
|
||||||
| NC score | 0.053912 (rank : 27) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86VB7, Q07898, Q07899, Q07900, Q07901, Q2VLH7 | Gene names | CD163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) (Hemoglobin scavenger receptor) [Contains: Soluble CD163 (sCD163)]. | |||||
|
C163A_MOUSE
|
||||||
| NC score | 0.053726 (rank : 28) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q2VLH6, Q2VLH5, Q99MX8 | Gene names | Cd163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) [Contains: Soluble CD163 (sCD163)]. | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.052320 (rank : 29) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.026416 (rank : 30) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
PLAC1_MOUSE
|
||||||
| NC score | 0.024768 (rank : 31) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JI83, Q3UKT4, Q80WW3 | Gene names | Plac1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Placenta-specific protein 1 precursor (EPCS26). | |||||
|
XKR5_HUMAN
|
||||||
| NC score | 0.019374 (rank : 32) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UX68, Q5GH74 | Gene names | XKR5, XRG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 5. | |||||
|
IGHA1_HUMAN
|
||||||
| NC score | 0.018213 (rank : 33) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01876 | Gene names | IGHA1 | |||
|
Domain Architecture |

|
|||||
| Description | Ig alpha-1 chain C region. | |||||
|
ZN750_HUMAN
|
||||||
| NC score | 0.014309 (rank : 34) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q32MQ0, Q9H899 | Gene names | ZNF750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
KCNH2_MOUSE
|
||||||
| NC score | 0.014285 (rank : 35) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
KCNH2_HUMAN
|
||||||
| NC score | 0.014278 (rank : 36) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.013744 (rank : 37) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.013087 (rank : 38) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
KR105_HUMAN
|
||||||
| NC score | 0.011828 (rank : 39) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
IF_HUMAN
|
||||||
| NC score | 0.011364 (rank : 40) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27352 | Gene names | GIF, IFMH | |||
|
Domain Architecture |

|
|||||
| Description | Gastric intrinsic factor precursor (Intrinsic factor) (IF) (INF). | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.010938 (rank : 41) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
ZN295_HUMAN
|
||||||
| NC score | 0.010755 (rank : 42) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULJ3, Q6P4R0 | Gene names | ZNF295, KIAA1227, ZBTB21 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 295 (Zinc finger and BTB domain-containing protein 21). | |||||
|
CAH9_MOUSE
|
||||||
| NC score | 0.010598 (rank : 43) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHB5, Q8K1G1, Q8VDE4 | Gene names | Ca9, Car9 | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase 9 precursor (EC 4.2.1.1) (Carbonic anhydrase IX) (Carbonate dehydratase IX) (CA-IX) (CAIX) (Membrane antigen MN homolog). | |||||
|
TSP1_MOUSE
|
||||||
| NC score | 0.009492 (rank : 44) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35441 | Gene names | Thbs1, Tsp1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
SH3K1_MOUSE
|
||||||
| NC score | 0.007946 (rank : 45) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R550, Q8CAL8, Q8CEF6, Q8R545, Q8R546, Q8R547, Q8R548, Q8R549, Q8R551, Q9CTQ9, Q9DC14, Q9JKC3 | Gene names | Sh3kbp1, Ruk, Seta | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (SH3-containing, expressed in tumorigenic astrocytes) (Regulator of ubiquitous kinase) (Ruk). | |||||
|
EMX2_HUMAN
|
||||||
| NC score | 0.006479 (rank : 46) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04743, Q96NN8, Q9BQF4 | Gene names | EMX2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein EMX2 (Empty spiracles homolog 2) (Empty spiracles- like protein 2). | |||||
|
EMX2_MOUSE
|
||||||
| NC score | 0.006465 (rank : 47) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04744, Q80UE7 | Gene names | Emx2, Emx-2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein EMX2 (Empty spiracles homolog 2) (Empty spiracles- like protein 2). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.003748 (rank : 48) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.002890 (rank : 49) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.002563 (rank : 50) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||