Please be patient as the page loads
|
TCEA3_MOUSE
|
||||||
| SwissProt Accessions | P23881, O88710, Q9CTZ8, Q9DCZ5 | Gene names | Tcea3, Tfiish | |||
|
Domain Architecture |
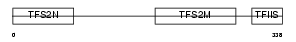
|
|||||
| Description | Transcription elongation factor A protein 3 (Transcription elongation factor S-II protein 3) (Transcription elongation factor TFIIS.h). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TCEA3_MOUSE
|
||||||
| θ value | 7.25522e-167 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P23881, O88710, Q9CTZ8, Q9DCZ5 | Gene names | Tcea3, Tfiish | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor A protein 3 (Transcription elongation factor S-II protein 3) (Transcription elongation factor TFIIS.h). | |||||
|
TCEA3_HUMAN
|
||||||
| θ value | 2.34478e-149 (rank : 2) | NC score | 0.992787 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75764 | Gene names | TCEA3, TFIISH | |||
|
Domain Architecture |
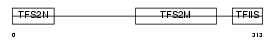
|
|||||
| Description | Transcription elongation factor A protein 3 (Transcription elongation factor S-II protein 3) (Transcription elongation factor TFIIS.h). | |||||
|
TCEA1_HUMAN
|
||||||
| θ value | 4.80189e-94 (rank : 3) | NC score | 0.972658 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P23193, Q15563, Q6FG87 | Gene names | TCEA1, TFIIS | |||
|
Domain Architecture |
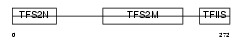
|
|||||
| Description | Transcription elongation factor A protein 1 (Transcription elongation factor S-II protein 1) (Transcription elongation factor TFIIS.o). | |||||
|
TCEA2_HUMAN
|
||||||
| θ value | 4.06505e-93 (rank : 4) | NC score | 0.971445 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15560, Q8TD37, Q8TD38 | Gene names | TCEA2 | |||
|
Domain Architecture |
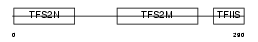
|
|||||
| Description | Transcription elongation factor A protein 2 (Transcription elongation factor S-II protein 2) (Testis-specific S-II) (Transcription elongation factor TFIIS.l). | |||||
|
TCEA1_MOUSE
|
||||||
| θ value | 7.17548e-90 (rank : 5) | NC score | 0.975344 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P10711, P10712, P23713 | Gene names | Tcea1, Tceat | |||
|
Domain Architecture |
|
|||||
| Description | Transcription elongation factor A protein 1 (Transcription elongation factor S-II protein 1) (Transcription elongation factor TFIIS.o). | |||||
|
TCEA2_MOUSE
|
||||||
| θ value | 2.08775e-89 (rank : 6) | NC score | 0.976335 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QVN7, O08667 | Gene names | Tcea2 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription elongation factor A protein 2 (Transcription elongation factor S-II protein 2) (Testis-specific S-II) (Protein S-II-T1) (Transcription elongation factor TFIIS.l). | |||||
|
PHF3_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 7) | NC score | 0.430109 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 8) | NC score | 0.230363 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
RPC11_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 9) | NC score | 0.477086 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y2Y1, Q96S35 | Gene names | POLR3K, RPC11 | |||
|
Domain Architecture |
|
|||||
| Description | DNA-directed RNA polymerases III 12.5 kDa polypeptide (EC 2.7.7.6) (RNA polymerase III subunit K) (RNA polymerase III C11 subunit) (HsC11p) (hRPC11). | |||||
|
RPC11_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 10) | NC score | 0.468697 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CQZ7, Q9D1U1 | Gene names | Polr3k | |||
|
Domain Architecture |
|
|||||
| Description | DNA-directed RNA polymerases III 12.5 kDa polypeptide (EC 2.7.7.6) (RNA polymerase III subunit K) (RNA polymerase III C11 subunit). | |||||
|
CRSP7_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 11) | NC score | 0.382340 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 12) | NC score | 0.219833 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
ELOA1_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 13) | NC score | 0.235794 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CB77, Q80VB2, Q9R0Q5 | Gene names | Tceb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 14) | NC score | 0.056416 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 15) | NC score | 0.061796 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
RPA12_HUMAN
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.148793 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P1U0 | Gene names | ZNRD1, RPA12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase I subunit 12 (EC 2.7.7.6) (Nuclear RNA polymerase I small-specific subunit Rpa12) (Zinc ribbon domain- containing protein 1). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.071998 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.060764 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
ZN383_HUMAN
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.010395 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NA42, Q6X2C7 | Gene names | ZNF383 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 383. | |||||
|
MBB1A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.032091 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
RPA12_MOUSE
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.123194 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q791N7 | Gene names | Znrd1, Rpa12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase I subunit 12 (EC 2.7.7.6) (Nuclear RNA polymerase I small-specific subunit Rpa12) (Zinc ribbon domain- containing protein 1). | |||||
|
APXL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.023311 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
ATRIP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.021877 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BMG1 | Gene names | Atrip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATR-interacting protein (ATM and Rad3-related-interacting protein). | |||||
|
CEP72_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.017956 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P209, Q9BV03, Q9BWM3, Q9NVR4 | Gene names | CEP72, KIAA1519 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 72 kDa (Cep72 protein). | |||||
|
HEAT3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.022486 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z4Q2, Q8N525, Q8WV56, Q96CC9, Q9NWN7 | Gene names | HEATR3 | |||
|
Domain Architecture |
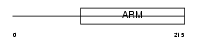
|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
PHF20_HUMAN
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.047837 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
PRGC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.018995 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70343 | Gene names | Ppargc1a, Pgc1, Pgc1a, Ppargc1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPAR gamma coactivator 1-alpha) (PPARGC-1-alpha) (PGC-1-alpha). | |||||
|
ZN157_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.005549 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51786, Q96LE9 | Gene names | ZNF157 | |||
|
Domain Architecture |
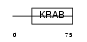
|
|||||
| Description | Zinc finger protein 157 (Zinc finger protein HZF22). | |||||
|
ZNF12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.010079 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P17014, Q9ULZ6 | Gene names | ZNF12, GIOT3, KOX3, ZNF325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 12 (Zinc finger protein KOX3) (Zinc finger protein 325) (Gonadotropin-inducible transcription repressor 3) (GIOT-3). | |||||
|
DNMT1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.019707 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P13864, P97413, Q80ZU3, Q9CSC6, Q9QXX6 | Gene names | Dnmt1, Dnmt, Met1, Uim | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase MmuI) (DNA MTase MmuI) (MCMT) (M.MmuI) (Met-1). | |||||
|
MAP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.019506 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.003368 (rank : 51) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.018261 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
U3IP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.007165 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43818, Q8IZ30 | Gene names | RNU3IP2, U355K | |||
|
Domain Architecture |

|
|||||
| Description | U3 small nucleolar RNA-interacting protein 2 (U3 small nucleolar ribonucleoprotein-associated 55 kDa protein) (U3 snoRNP-associated 55 kDa protein) (U3-55K). | |||||
|
C8AP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.019514 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.010073 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
EVL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.012185 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.008302 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
PP1RA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.113889 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96QC0, O00405 | Gene names | PPP1R10, CAT53, FB19, PNUTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (Phosphatase 1 nuclear targeting subunit) (MHC class I region proline- rich protein CAT53) (FB19 protein) (PP1-binding protein of 114 kDa) (p99). | |||||
|
PP1RA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.110053 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80W00, Q811B6, Q8C6T7, Q8K2U8 | Gene names | Ppp1r10, Cat53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (MHC class I region proline-rich protein CAT53). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.015646 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
NPHP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.007147 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15259, O14837 | Gene names | NPHP1, NPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Nephrocystin-1 (Juvenile nephronophthisis 1 protein). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.004993 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
PHLPP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.005380 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.032517 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.015560 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
ZN436_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.001191 (rank : 52) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BPP0, Q69ZC6, Q8BQC2 | Gene names | Znf436, Kiaa1710, Zfp46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 436 (Zinc finger protein 46). | |||||
|
ZN570_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.005032 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96NI8 | Gene names | ZNF570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 570. | |||||
|
ELOA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.111109 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IYF1, Q9P2V9 | Gene names | TCEB3B, TCEB3L | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A2 (Elongin-A2) (EloA2) (Transcription elongation factor B polypeptide 3B). | |||||
|
ELOA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.129006 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NG57 | Gene names | TCEB3C, TCEB3L2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A3 (Elongin-A3) (EloA3) (Transcription elongation factor B polypeptide 3C). | |||||
|
RPB9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.096910 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P36954, Q6NW05 | Gene names | POLR2I | |||
|
Domain Architecture |
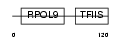
|
|||||
| Description | DNA-directed RNA polymerase II subunit I (EC 2.7.7.6) (DNA-directed RNA polymerase II 14.5 kDa polypeptide) (RPB9) (RPB14.5). | |||||
|
RPB9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.096910 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P60898 | Gene names | Polr2i | |||
|
Domain Architecture |
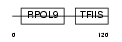
|
|||||
| Description | DNA-directed RNA polymerase II subunit I (EC 2.7.7.6) (DNA-directed RNA polymerase II 14.5 kDa polypeptide) (RPB9) (RPB14.5). | |||||
|
TCEA3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 7.25522e-167 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P23881, O88710, Q9CTZ8, Q9DCZ5 | Gene names | Tcea3, Tfiish | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor A protein 3 (Transcription elongation factor S-II protein 3) (Transcription elongation factor TFIIS.h). | |||||
|
TCEA3_HUMAN
|
||||||
| NC score | 0.992787 (rank : 2) | θ value | 2.34478e-149 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75764 | Gene names | TCEA3, TFIISH | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor A protein 3 (Transcription elongation factor S-II protein 3) (Transcription elongation factor TFIIS.h). | |||||
|
TCEA2_MOUSE
|
||||||
| NC score | 0.976335 (rank : 3) | θ value | 2.08775e-89 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QVN7, O08667 | Gene names | Tcea2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor A protein 2 (Transcription elongation factor S-II protein 2) (Testis-specific S-II) (Protein S-II-T1) (Transcription elongation factor TFIIS.l). | |||||
|
TCEA1_MOUSE
|
||||||
| NC score | 0.975344 (rank : 4) | θ value | 7.17548e-90 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P10711, P10712, P23713 | Gene names | Tcea1, Tceat | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor A protein 1 (Transcription elongation factor S-II protein 1) (Transcription elongation factor TFIIS.o). | |||||
|
TCEA1_HUMAN
|
||||||
| NC score | 0.972658 (rank : 5) | θ value | 4.80189e-94 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P23193, Q15563, Q6FG87 | Gene names | TCEA1, TFIIS | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor A protein 1 (Transcription elongation factor S-II protein 1) (Transcription elongation factor TFIIS.o). | |||||
|
TCEA2_HUMAN
|
||||||
| NC score | 0.971445 (rank : 6) | θ value | 4.06505e-93 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15560, Q8TD37, Q8TD38 | Gene names | TCEA2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor A protein 2 (Transcription elongation factor S-II protein 2) (Testis-specific S-II) (Transcription elongation factor TFIIS.l). | |||||
|
RPC11_HUMAN
|
||||||
| NC score | 0.477086 (rank : 7) | θ value | 1.43324e-05 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y2Y1, Q96S35 | Gene names | POLR3K, RPC11 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerases III 12.5 kDa polypeptide (EC 2.7.7.6) (RNA polymerase III subunit K) (RNA polymerase III C11 subunit) (HsC11p) (hRPC11). | |||||
|
RPC11_MOUSE
|
||||||
| NC score | 0.468697 (rank : 8) | θ value | 1.43324e-05 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CQZ7, Q9D1U1 | Gene names | Polr3k | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerases III 12.5 kDa polypeptide (EC 2.7.7.6) (RNA polymerase III subunit K) (RNA polymerase III C11 subunit). | |||||
|
PHF3_HUMAN
|
||||||
| NC score | 0.430109 (rank : 9) | θ value | 1.33837e-11 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
CRSP7_HUMAN
|
||||||
| NC score | 0.382340 (rank : 10) | θ value | 4.1701e-05 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
ELOA1_MOUSE
|
||||||
| NC score | 0.235794 (rank : 11) | θ value | 0.00102713 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CB77, Q80VB2, Q9R0Q5 | Gene names | Tceb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.230363 (rank : 12) | θ value | 1.69304e-06 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.219833 (rank : 13) | θ value | 0.00020696 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
RPA12_HUMAN
|
||||||
| NC score | 0.148793 (rank : 14) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P1U0 | Gene names | ZNRD1, RPA12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase I subunit 12 (EC 2.7.7.6) (Nuclear RNA polymerase I small-specific subunit Rpa12) (Zinc ribbon domain- containing protein 1). | |||||
|
ELOA3_HUMAN
|
||||||
| NC score | 0.129006 (rank : 15) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NG57 | Gene names | TCEB3C, TCEB3L2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A3 (Elongin-A3) (EloA3) (Transcription elongation factor B polypeptide 3C). | |||||
|
RPA12_MOUSE
|
||||||
| NC score | 0.123194 (rank : 16) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q791N7 | Gene names | Znrd1, Rpa12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase I subunit 12 (EC 2.7.7.6) (Nuclear RNA polymerase I small-specific subunit Rpa12) (Zinc ribbon domain- containing protein 1). | |||||
|
PP1RA_HUMAN
|
||||||
| NC score | 0.113889 (rank : 17) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96QC0, O00405 | Gene names | PPP1R10, CAT53, FB19, PNUTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (Phosphatase 1 nuclear targeting subunit) (MHC class I region proline- rich protein CAT53) (FB19 protein) (PP1-binding protein of 114 kDa) (p99). | |||||
|
ELOA2_HUMAN
|
||||||
| NC score | 0.111109 (rank : 18) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IYF1, Q9P2V9 | Gene names | TCEB3B, TCEB3L | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A2 (Elongin-A2) (EloA2) (Transcription elongation factor B polypeptide 3B). | |||||
|
PP1RA_MOUSE
|
||||||
| NC score | 0.110053 (rank : 19) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80W00, Q811B6, Q8C6T7, Q8K2U8 | Gene names | Ppp1r10, Cat53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (MHC class I region proline-rich protein CAT53). | |||||
|
RPB9_HUMAN
|
||||||
| NC score | 0.096910 (rank : 20) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P36954, Q6NW05 | Gene names | POLR2I | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II subunit I (EC 2.7.7.6) (DNA-directed RNA polymerase II 14.5 kDa polypeptide) (RPB9) (RPB14.5). | |||||
|
RPB9_MOUSE
|
||||||
| NC score | 0.096910 (rank : 21) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P60898 | Gene names | Polr2i | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II subunit I (EC 2.7.7.6) (DNA-directed RNA polymerase II 14.5 kDa polypeptide) (RPB9) (RPB14.5). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.071998 (rank : 22) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.061796 (rank : 23) | θ value | 0.0252991 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.060764 (rank : 24) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.056416 (rank : 25) | θ value | 0.0193708 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PHF20_HUMAN
|
||||||
| NC score | 0.047837 (rank : 26) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.032517 (rank : 27) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
MBB1A_MOUSE
|
||||||
| NC score | 0.032091 (rank : 28) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.023311 (rank : 29) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
HEAT3_HUMAN
|
||||||
| NC score | 0.022486 (rank : 30) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z4Q2, Q8N525, Q8WV56, Q96CC9, Q9NWN7 | Gene names | HEATR3 | |||
|
Domain Architecture |

|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
ATRIP_MOUSE
|
||||||
| NC score | 0.021877 (rank : 31) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BMG1 | Gene names | Atrip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATR-interacting protein (ATM and Rad3-related-interacting protein). | |||||
|
DNMT1_MOUSE
|
||||||
| NC score | 0.019707 (rank : 32) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P13864, P97413, Q80ZU3, Q9CSC6, Q9QXX6 | Gene names | Dnmt1, Dnmt, Met1, Uim | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase MmuI) (DNA MTase MmuI) (MCMT) (M.MmuI) (Met-1). | |||||
|
C8AP2_HUMAN
|
||||||
| NC score | 0.019514 (rank : 33) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
MAP2_MOUSE
|
||||||
| NC score | 0.019506 (rank : 34) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
PRGC1_MOUSE
|
||||||
| NC score | 0.018995 (rank : 35) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70343 | Gene names | Ppargc1a, Pgc1, Pgc1a, Ppargc1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPAR gamma coactivator 1-alpha) (PPARGC-1-alpha) (PGC-1-alpha). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.018261 (rank : 36) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CEP72_HUMAN
|
||||||
| NC score | 0.017956 (rank : 37) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P209, Q9BV03, Q9BWM3, Q9NVR4 | Gene names | CEP72, KIAA1519 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 72 kDa (Cep72 protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.015646 (rank : 38) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.015560 (rank : 39) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
EVL_HUMAN
|
||||||
| NC score | 0.012185 (rank : 40) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
ZN383_HUMAN
|
||||||
| NC score | 0.010395 (rank : 41) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NA42, Q6X2C7 | Gene names | ZNF383 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 383. | |||||
|
ZNF12_HUMAN
|
||||||
| NC score | 0.010079 (rank : 42) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P17014, Q9ULZ6 | Gene names | ZNF12, GIOT3, KOX3, ZNF325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 12 (Zinc finger protein KOX3) (Zinc finger protein 325) (Gonadotropin-inducible transcription repressor 3) (GIOT-3). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.010073 (rank : 43) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.008302 (rank : 44) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
U3IP2_HUMAN
|
||||||
| NC score | 0.007165 (rank : 45) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43818, Q8IZ30 | Gene names | RNU3IP2, U355K | |||
|
Domain Architecture |

|
|||||
| Description | U3 small nucleolar RNA-interacting protein 2 (U3 small nucleolar ribonucleoprotein-associated 55 kDa protein) (U3 snoRNP-associated 55 kDa protein) (U3-55K). | |||||
|
NPHP1_HUMAN
|
||||||
| NC score | 0.007147 (rank : 46) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15259, O14837 | Gene names | NPHP1, NPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Nephrocystin-1 (Juvenile nephronophthisis 1 protein). | |||||
|
ZN157_HUMAN
|
||||||
| NC score | 0.005549 (rank : 47) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51786, Q96LE9 | Gene names | ZNF157 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 157 (Zinc finger protein HZF22). | |||||
|
PHLPP_MOUSE
|
||||||
| NC score | 0.005380 (rank : 48) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHE4, Q6PCN0, Q8QZU8, Q8R5E5, Q99KL6 | Gene names | Phlpp, Kiaa0606, Plekhe1, Scop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein). | |||||
|
ZN570_HUMAN
|
||||||
| NC score | 0.005032 (rank : 49) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96NI8 | Gene names | ZNF570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 570. | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.004993 (rank : 50) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.003368 (rank : 51) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
ZN436_MOUSE
|
||||||
| NC score | 0.001191 (rank : 52) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BPP0, Q69ZC6, Q8BQC2 | Gene names | Znf436, Kiaa1710, Zfp46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 436 (Zinc finger protein 46). | |||||