Please be patient as the page loads
|
STAP1_HUMAN
|
||||||
| SwissProt Accessions | Q9ULZ2 | Gene names | STAP1, BRDG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 1 (STAP-1) (Stem cell adaptor protein 1) (BCR downstream signaling protein 1) (Docking protein BRDG1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
STAP1_HUMAN
|
||||||
| θ value | 4.86655e-163 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ULZ2 | Gene names | STAP1, BRDG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 1 (STAP-1) (Stem cell adaptor protein 1) (BCR downstream signaling protein 1) (Docking protein BRDG1). | |||||
|
STAP1_MOUSE
|
||||||
| θ value | 1.12976e-135 (rank : 2) | NC score | 0.989896 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JM90, Q3TRM1, Q6PES6 | Gene names | Stap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 1 (STAP-1) (Stem cell adaptor protein 1). | |||||
|
STAP2_MOUSE
|
||||||
| θ value | 1.28434e-38 (rank : 3) | NC score | 0.836040 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R0L1, Q8BWS2 | Gene names | Stap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 2 (STAP-2). | |||||
|
STAP2_HUMAN
|
||||||
| θ value | 4.13158e-37 (rank : 4) | NC score | 0.839431 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UGK3, Q9NXI2 | Gene names | STAP2, BKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 2 (STAP-2) (Breast tumor kinase substrate) (BRK substrate). | |||||
|
DGKH_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 5) | NC score | 0.145315 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86XP1, Q5VZW0, Q6PI56, Q86XP2, Q8N3N0, Q8N7J9 | Gene names | DGKH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase eta (EC 2.7.1.107) (Diglyceride kinase eta) (DGK-eta) (DAG kinase eta). | |||||
|
DGKD_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 6) | NC score | 0.134757 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16760, Q14158, Q6PK55, Q8NG53 | Gene names | DGKD, KIAA0145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase delta (EC 2.7.1.107) (Diglyceride kinase delta) (DGK-delta) (DAG kinase delta) (130 kDa diacylglycerol kinase). | |||||
|
SRMS_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 7) | NC score | 0.012857 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H3Y6 | Gene names | SRMS, C20orf148 | |||
|
Domain Architecture |
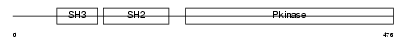
|
|||||
| Description | Tyrosine-protein kinase Srms (EC 2.7.10.2). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 8) | NC score | 0.029173 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
PLCG2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.051836 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16885, Q969T5 | Gene names | PLCG2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-2) (Phospholipase C-gamma-2) (PLC-IV). | |||||
|
TIAM1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 10) | NC score | 0.062032 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60610 | Gene names | Tiam1, Tiam-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
TOM1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 11) | NC score | 0.034666 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60784, Q5TIJ6 | Gene names | TOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TIAM1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 12) | NC score | 0.058084 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
TTC1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 13) | NC score | 0.035549 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
FINC_MOUSE
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.021279 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.008940 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
FGD5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.020020 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80UZ0, Q8BHM5 | Gene names | Fgd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.007218 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
TTC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.030128 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.006216 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
FINC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.018930 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
NKX61_MOUSE
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.005718 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MA9, Q9ERQ7 | Gene names | Nkx6-1, Nkx6.1, Nkx6a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
FGD6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.013975 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
DGKK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.060840 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
STAP1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.86655e-163 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ULZ2 | Gene names | STAP1, BRDG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 1 (STAP-1) (Stem cell adaptor protein 1) (BCR downstream signaling protein 1) (Docking protein BRDG1). | |||||
|
STAP1_MOUSE
|
||||||
| NC score | 0.989896 (rank : 2) | θ value | 1.12976e-135 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JM90, Q3TRM1, Q6PES6 | Gene names | Stap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 1 (STAP-1) (Stem cell adaptor protein 1). | |||||
|
STAP2_HUMAN
|
||||||
| NC score | 0.839431 (rank : 3) | θ value | 4.13158e-37 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UGK3, Q9NXI2 | Gene names | STAP2, BKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 2 (STAP-2) (Breast tumor kinase substrate) (BRK substrate). | |||||
|
STAP2_MOUSE
|
||||||
| NC score | 0.836040 (rank : 4) | θ value | 1.28434e-38 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R0L1, Q8BWS2 | Gene names | Stap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 2 (STAP-2). | |||||
|
DGKH_HUMAN
|
||||||
| NC score | 0.145315 (rank : 5) | θ value | 1.09739e-05 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86XP1, Q5VZW0, Q6PI56, Q86XP2, Q8N3N0, Q8N7J9 | Gene names | DGKH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase eta (EC 2.7.1.107) (Diglyceride kinase eta) (DGK-eta) (DAG kinase eta). | |||||
|
DGKD_HUMAN
|
||||||
| NC score | 0.134757 (rank : 6) | θ value | 0.00020696 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16760, Q14158, Q6PK55, Q8NG53 | Gene names | DGKD, KIAA0145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase delta (EC 2.7.1.107) (Diglyceride kinase delta) (DGK-delta) (DAG kinase delta) (130 kDa diacylglycerol kinase). | |||||
|
TIAM1_MOUSE
|
||||||
| NC score | 0.062032 (rank : 7) | θ value | 0.365318 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60610 | Gene names | Tiam1, Tiam-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
DGKK_HUMAN
|
||||||
| NC score | 0.060840 (rank : 8) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
TIAM1_HUMAN
|
||||||
| NC score | 0.058084 (rank : 9) | θ value | 0.813845 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13009 | Gene names | TIAM1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
PLCG2_HUMAN
|
||||||
| NC score | 0.051836 (rank : 10) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16885, Q969T5 | Gene names | PLCG2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-2) (Phospholipase C-gamma-2) (PLC-IV). | |||||
|
TTC1_MOUSE
|
||||||
| NC score | 0.035549 (rank : 11) | θ value | 1.06291 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
TOM1_HUMAN
|
||||||
| NC score | 0.034666 (rank : 12) | θ value | 0.47712 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60784, Q5TIJ6 | Gene names | TOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TTC1_HUMAN
|
||||||
| NC score | 0.030128 (rank : 13) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99614, Q9BVT3 | Gene names | TTC1, TPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.029173 (rank : 14) | θ value | 0.125558 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
FINC_MOUSE
|
||||||
| NC score | 0.021279 (rank : 15) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
FGD5_MOUSE
|
||||||
| NC score | 0.020020 (rank : 16) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80UZ0, Q8BHM5 | Gene names | Fgd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 5. | |||||
|
FINC_HUMAN
|
||||||
| NC score | 0.018930 (rank : 17) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
FGD6_HUMAN
|
||||||
| NC score | 0.013975 (rank : 18) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
SRMS_HUMAN
|
||||||
| NC score | 0.012857 (rank : 19) | θ value | 0.0736092 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H3Y6 | Gene names | SRMS, C20orf148 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Srms (EC 2.7.10.2). | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.008940 (rank : 20) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.007218 (rank : 21) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.006216 (rank : 22) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
NKX61_MOUSE
|
||||||
| NC score | 0.005718 (rank : 23) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MA9, Q9ERQ7 | Gene names | Nkx6-1, Nkx6.1, Nkx6a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||