Please be patient as the page loads
|
SARDH_HUMAN
|
||||||
| SwissProt Accessions | Q9UL12, Q9Y280, Q9Y2Y3 | Gene names | SARDH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcosine dehydrogenase, mitochondrial precursor (EC 1.5.99.1) (SarDH) (BPR-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SARDH_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UL12, Q9Y280, Q9Y2Y3 | Gene names | SARDH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcosine dehydrogenase, mitochondrial precursor (EC 1.5.99.1) (SarDH) (BPR-2). | |||||
|
SARDH_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.996500 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99LB7 | Gene names | Sardh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcosine dehydrogenase, mitochondrial precursor (EC 1.5.99.1) (SarDH). | |||||
|
M2GD_HUMAN
|
||||||
| θ value | 3.90098e-120 (rank : 3) | NC score | 0.913934 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UI17 | Gene names | DMGDH | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
M2GD_MOUSE
|
||||||
| θ value | 2.79562e-118 (rank : 4) | NC score | 0.911602 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9DBT9, Q8R1S7 | Gene names | Dmgdh | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
GCST_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 5) | NC score | 0.525442 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48728 | Gene names | AMT, GCST | |||
|
Domain Architecture |
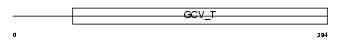
|
|||||
| Description | Aminomethyltransferase, mitochondrial precursor (EC 2.1.2.10) (Glycine cleavage system T protein) (GCVT). | |||||
|
GCST_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 6) | NC score | 0.499817 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CFA2 | Gene names | Amt | |||
|
Domain Architecture |
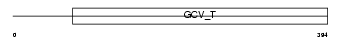
|
|||||
| Description | Aminomethyltransferase, mitochondrial precursor (EC 2.1.2.10) (Glycine cleavage system T protein) (GCVT). | |||||
|
L2HDH_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 7) | NC score | 0.194062 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H9P8, Q9BRR1 | Gene names | L2HGDH, C14orf160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-2-hydroxyglutarate dehydrogenase, mitochondrial precursor (EC 1.1.99.2) (Duranin). | |||||
|
GPDM_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 8) | NC score | 0.181725 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64521, Q61507 | Gene names | Gpd2, Gdm1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M). | |||||
|
L2HDH_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 9) | NC score | 0.163955 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91YP0, Q3TH61, Q3U7Z0, Q3ULY6 | Gene names | L2hgdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-2-hydroxyglutarate dehydrogenase, mitochondrial precursor (EC 1.1.99.2) (Duranin). | |||||
|
SOX_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 10) | NC score | 0.270397 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P0Z9, Q96H28, Q9C070 | Gene names | PIPOX, LPIPOX, PSO | |||
|
Domain Architecture |
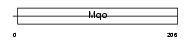
|
|||||
| Description | Peroxisomal sarcosine oxidase (EC 1.5.3.1) (PSO) (L-pipecolate oxidase) (EC 1.5.3.7) (L-pipecolic acid oxidase). | |||||
|
SOX_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.206096 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D826, O55223 | Gene names | Pipox, Pso | |||
|
Domain Architecture |
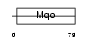
|
|||||
| Description | Peroxisomal sarcosine oxidase (EC 1.5.3.1) (PSO) (L-pipecolate oxidase) (EC 1.5.3.7) (L-pipecolic acid oxidase). | |||||
|
INADL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 12) | NC score | 0.011269 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.018503 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
AATK_MOUSE
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.003490 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80YE4, O35211, Q3U2U5, Q3UHR8, Q66JN3, Q80YE3, Q8CB63, Q8CHE2 | Gene names | Aatk, Kiaa0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase). | |||||
|
DLDH_MOUSE
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.038606 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08749 | Gene names | Dld | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.018186 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.015727 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
MSLN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.030857 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13421, Q14859, Q4VQD5, Q96GR6, Q96KJ5, Q9BR17, Q9BTR2, Q9UK57 | Gene names | MSLN, MPF | |||
|
Domain Architecture |

|
|||||
| Description | Mesothelin precursor (Pre-pro-megakaryocyte potentiating factor) (CAK1 antigen) [Contains: Megakaryocyte-potentiating factor (MPF); Mesothelin, cleaved form]. | |||||
|
MCPH1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.013558 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
OXDA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.013065 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P14920, Q16758, Q8N6R2 | Gene names | DAO, DAMOX | |||
|
Domain Architecture |
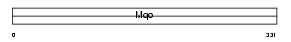
|
|||||
| Description | D-amino-acid oxidase (EC 1.4.3.3) (DAMOX) (DAO) (DAAO). | |||||
|
SIA7D_HUMAN
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.014291 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H4F1, Q9NWU6, Q9UKU1, Q9ULB9, Q9Y3G3, Q9Y3G4 | Gene names | ST6GALNAC4, SIAT3C, SIAT7D | |||
|
Domain Architecture |
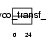
|
|||||
| Description | Alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3-N-acetyl- galactosaminide alpha-2,6-sialyltransferase (EC 2.4.99.7) (NeuAc- alpha-2,3-Gal-beta-1,3-GalNAc-alpha-2,6-sialyltransferase) (ST6GalNAc IV) (Sialyltransferase 7D) (Sialyltransferase 3C). | |||||
|
MMP25_HUMAN
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.003720 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPA2, Q9H3Q0 | Gene names | MMP25, MT6MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-25 precursor (EC 3.4.24.-) (MMP-25) (Membrane-type matrix metalloproteinase 6) (MT-MMP 6) (Membrane-type-6 matrix metalloproteinase) (MT6-MMP) (Leukolysin). | |||||
|
GPDM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.124754 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P43304, Q59FR1, Q9HAP9 | Gene names | GPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M) (mtGPD). | |||||
|
SARDH_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UL12, Q9Y280, Q9Y2Y3 | Gene names | SARDH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcosine dehydrogenase, mitochondrial precursor (EC 1.5.99.1) (SarDH) (BPR-2). | |||||
|
SARDH_MOUSE
|
||||||
| NC score | 0.996500 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99LB7 | Gene names | Sardh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcosine dehydrogenase, mitochondrial precursor (EC 1.5.99.1) (SarDH). | |||||
|
M2GD_HUMAN
|
||||||
| NC score | 0.913934 (rank : 3) | θ value | 3.90098e-120 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UI17 | Gene names | DMGDH | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
M2GD_MOUSE
|
||||||
| NC score | 0.911602 (rank : 4) | θ value | 2.79562e-118 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9DBT9, Q8R1S7 | Gene names | Dmgdh | |||
|
Domain Architecture |

|
|||||
| Description | Dimethylglycine dehydrogenase, mitochondrial precursor (EC 1.5.99.2) (ME2GLYDH). | |||||
|
GCST_HUMAN
|
||||||
| NC score | 0.525442 (rank : 5) | θ value | 4.59992e-12 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48728 | Gene names | AMT, GCST | |||
|
Domain Architecture |

|
|||||
| Description | Aminomethyltransferase, mitochondrial precursor (EC 2.1.2.10) (Glycine cleavage system T protein) (GCVT). | |||||
|
GCST_MOUSE
|
||||||
| NC score | 0.499817 (rank : 6) | θ value | 3.64472e-09 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CFA2 | Gene names | Amt | |||
|
Domain Architecture |

|
|||||
| Description | Aminomethyltransferase, mitochondrial precursor (EC 2.1.2.10) (Glycine cleavage system T protein) (GCVT). | |||||
|
SOX_HUMAN
|
||||||
| NC score | 0.270397 (rank : 7) | θ value | 0.0148317 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P0Z9, Q96H28, Q9C070 | Gene names | PIPOX, LPIPOX, PSO | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal sarcosine oxidase (EC 1.5.3.1) (PSO) (L-pipecolate oxidase) (EC 1.5.3.7) (L-pipecolic acid oxidase). | |||||
|
SOX_MOUSE
|
||||||
| NC score | 0.206096 (rank : 8) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D826, O55223 | Gene names | Pipox, Pso | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal sarcosine oxidase (EC 1.5.3.1) (PSO) (L-pipecolate oxidase) (EC 1.5.3.7) (L-pipecolic acid oxidase). | |||||
|
L2HDH_HUMAN
|
||||||
| NC score | 0.194062 (rank : 9) | θ value | 5.44631e-05 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H9P8, Q9BRR1 | Gene names | L2HGDH, C14orf160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-2-hydroxyglutarate dehydrogenase, mitochondrial precursor (EC 1.1.99.2) (Duranin). | |||||
|
GPDM_MOUSE
|
||||||
| NC score | 0.181725 (rank : 10) | θ value | 9.29e-05 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64521, Q61507 | Gene names | Gpd2, Gdm1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M). | |||||
|
L2HDH_MOUSE
|
||||||
| NC score | 0.163955 (rank : 11) | θ value | 0.00390308 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91YP0, Q3TH61, Q3U7Z0, Q3ULY6 | Gene names | L2hgdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-2-hydroxyglutarate dehydrogenase, mitochondrial precursor (EC 1.1.99.2) (Duranin). | |||||
|
GPDM_HUMAN
|
||||||
| NC score | 0.124754 (rank : 12) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P43304, Q59FR1, Q9HAP9 | Gene names | GPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M) (mtGPD). | |||||
|
DLDH_MOUSE
|
||||||
| NC score | 0.038606 (rank : 13) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08749 | Gene names | Dld | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase). | |||||
|
MSLN_HUMAN
|
||||||
| NC score | 0.030857 (rank : 14) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13421, Q14859, Q4VQD5, Q96GR6, Q96KJ5, Q9BR17, Q9BTR2, Q9UK57 | Gene names | MSLN, MPF | |||
|
Domain Architecture |

|
|||||
| Description | Mesothelin precursor (Pre-pro-megakaryocyte potentiating factor) (CAK1 antigen) [Contains: Megakaryocyte-potentiating factor (MPF); Mesothelin, cleaved form]. | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.018503 (rank : 15) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.018186 (rank : 16) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.015727 (rank : 17) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
SIA7D_HUMAN
|
||||||
| NC score | 0.014291 (rank : 18) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H4F1, Q9NWU6, Q9UKU1, Q9ULB9, Q9Y3G3, Q9Y3G4 | Gene names | ST6GALNAC4, SIAT3C, SIAT7D | |||
|
Domain Architecture |
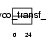
|
|||||
| Description | Alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3-N-acetyl- galactosaminide alpha-2,6-sialyltransferase (EC 2.4.99.7) (NeuAc- alpha-2,3-Gal-beta-1,3-GalNAc-alpha-2,6-sialyltransferase) (ST6GalNAc IV) (Sialyltransferase 7D) (Sialyltransferase 3C). | |||||
|
MCPH1_MOUSE
|
||||||
| NC score | 0.013558 (rank : 19) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
OXDA_HUMAN
|
||||||
| NC score | 0.013065 (rank : 20) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P14920, Q16758, Q8N6R2 | Gene names | DAO, DAMOX | |||
|
Domain Architecture |

|
|||||
| Description | D-amino-acid oxidase (EC 1.4.3.3) (DAMOX) (DAO) (DAAO). | |||||
|
INADL_HUMAN
|
||||||
| NC score | 0.011269 (rank : 21) | θ value | 1.81305 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
MMP25_HUMAN
|
||||||
| NC score | 0.003720 (rank : 22) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPA2, Q9H3Q0 | Gene names | MMP25, MT6MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-25 precursor (EC 3.4.24.-) (MMP-25) (Membrane-type matrix metalloproteinase 6) (MT-MMP 6) (Membrane-type-6 matrix metalloproteinase) (MT6-MMP) (Leukolysin). | |||||
|
AATK_MOUSE
|
||||||
| NC score | 0.003490 (rank : 23) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80YE4, O35211, Q3U2U5, Q3UHR8, Q66JN3, Q80YE3, Q8CB63, Q8CHE2 | Gene names | Aatk, Kiaa0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase). | |||||